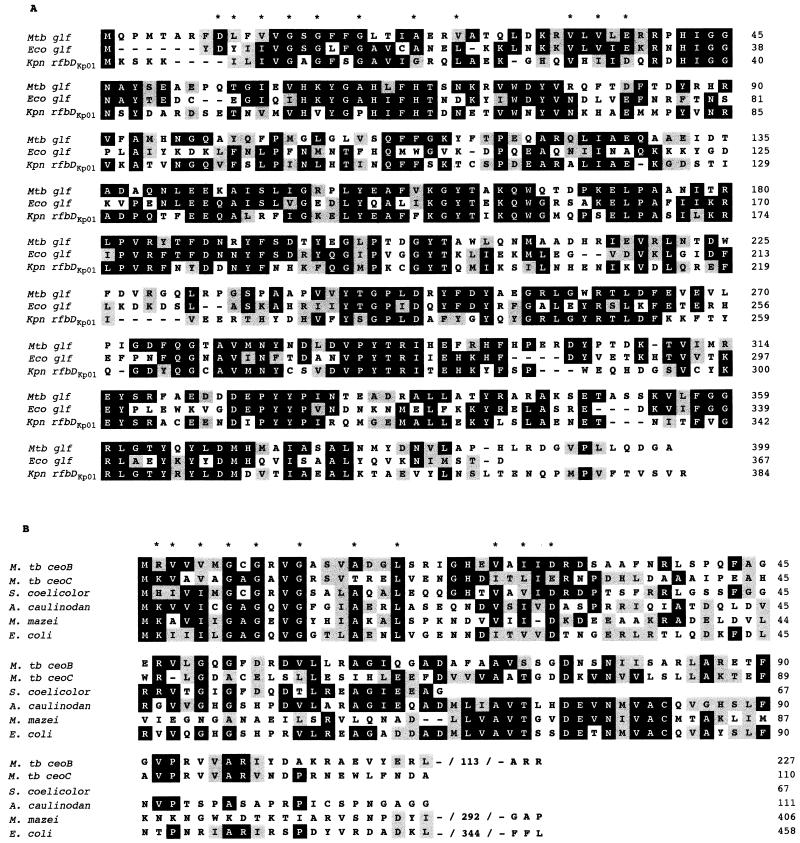
FIG. 2.
Amino acid sequence alignments among Glf and RfbD homologues (A) and among CeoB, CeoC, and TrkA homologues (B). Identical amino acid residues are grouped in black boxes, and conserved residues are indicated in gray. Those amino acids involved in the NAD+ binding motif are identified with asterisks. The alignment was performed by the method of Hein (10) and is available in the DNAStar software package.