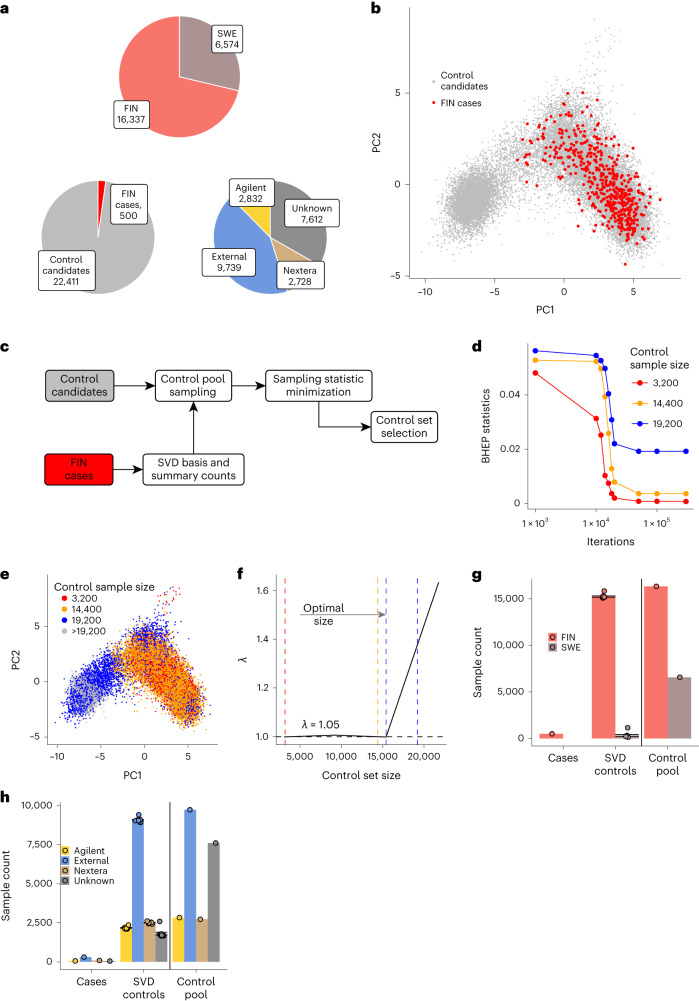
Fig. 3. Case study 2: simulated case–control study with 22,911 exomes dataset.
a, Breakdown of fine-scale ancestries, different exome platforms present in the dataset, and case–control study setup: random 500 Finnish samples are selected as ‘cases’ and the rest of the data is tested as prospective controls. b, Conventional PCA showing Finnish samples selected as a case group. c, Scheme of data handling simulating association study without genotype sharing. d, Sampling statistic minimization for control candidate sets of different size for the selected case cohort. e, Conventional PCA shows that greater size of the control candidate sets to be sampled deteriorates their quality, as can be seen by inclusion of samples of nontarget ancestry. f, Optimal size of the control pool for a given case cohort is selected to deliver the largest set of samples with λGC < 1.05. g, Matching experiment summary results over ten random sets of Finnish cases (error bars represent standard error; center of the error bars represents mean). h, Samples from multiple sequencing platforms are successfully selected as control group in ten random case sets (error bars represent standard error; center of the error bars represents mean). FIN, Finnish ancestry; SWE, Swedish ancestry.