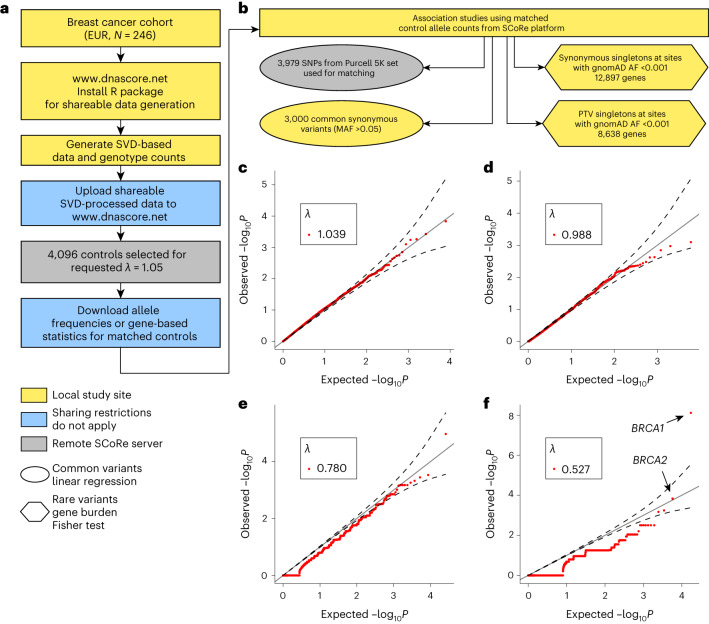
Fig. 5. Breast cancer samples (N = 244) matched to controls (N = 4,096) using SCoRe web service.
a, Workflow scheme for association study without genotype sharing. b, Control sets selected using different user-defined matching quality thresholds (λ). c, QQ plots for linear regression association statistics for every selection threshold on DNA variants used for matching. d, QQ plot for linear regression association statistics using summary genotypes counts from optimal control dataset (λ < 1.05) on common synonymous DNA variant. e, QQ plot for Fisher’s exact test association statistics using summary gene burden statistics for synonymous singletons on DNA variants with allele frequency <1 × 10−3 or not present in gnomAD. The solid line represents the diagonal, and the dashed lines indicate the 95% confidence interval (two-sided Fisher’s exact test). Raw, unadjusted P values are reported. f, QQ plot for Fisher’s exact test association statistics using summary gene burden statistics for protein-truncating singletons on DNA variants with allele frequency <1 × 10−3 or not present in gnomAD. The solid line represents the diagonal, and the dashed lines indicate the 95% confidence interval (two-sided Fisher’s exact test). Raw, unadjusted P values are reported. EUR, European and European-American ancestry; AF, allele frequency; SNP, single nucleotide polymorphism.