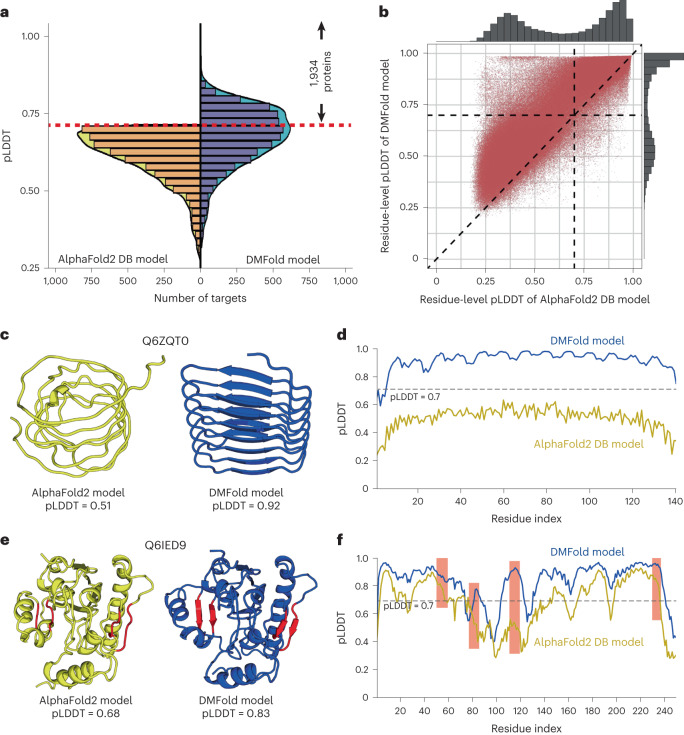
Fig. 4. The structural modeling results of DMFold on 5,042 difficult targets from the human proteome.
a, Distributions of pLDDTs for DMFold models versus AlphaFold2 DB models for the subset of 5,042 AlphaFold2 DB models with pLDDT < 0.70. The red dashed line marks the threshold pLDDT = 0.70 for considering a target to be confidently predicted, where DMFold models have a pLDDT ≥ 0.7 in 1,934 cases. b, Head-to-head comparison of the residue-level pLDDTs obtained by DMFold and AlphaFold2 DB for the 1,934 confidently DMFold-modeled proteins, which involve in total 878,094 residues. c, Structural models generated for a putative uncharacterized protein FLJ45035 (Q6ZQT0) by AlphaFold2 DB (yellow) and DMFold (blue), respectively. d, Residue-level pLDDT curves of AlphaFold2 DB (yellow) and DMFold (blue) for Q6ZQT0. e, Structural models generated for a putative diacylglycerol O-acyltransferase 2-like protein (Q6IED9) by AlphaFold2 DB (yellow) and DMFold (blue), respectively, where two better-formed β-sheet secondary structures created by DMFold are highlighted by red. f, Residue-level pLDDT curves of AlphaFold2 DB (yellow) and DMFold (blue) for Q6IED9, where the pLDDTs associated with the four β-strands are highlighted with red backgrounds.