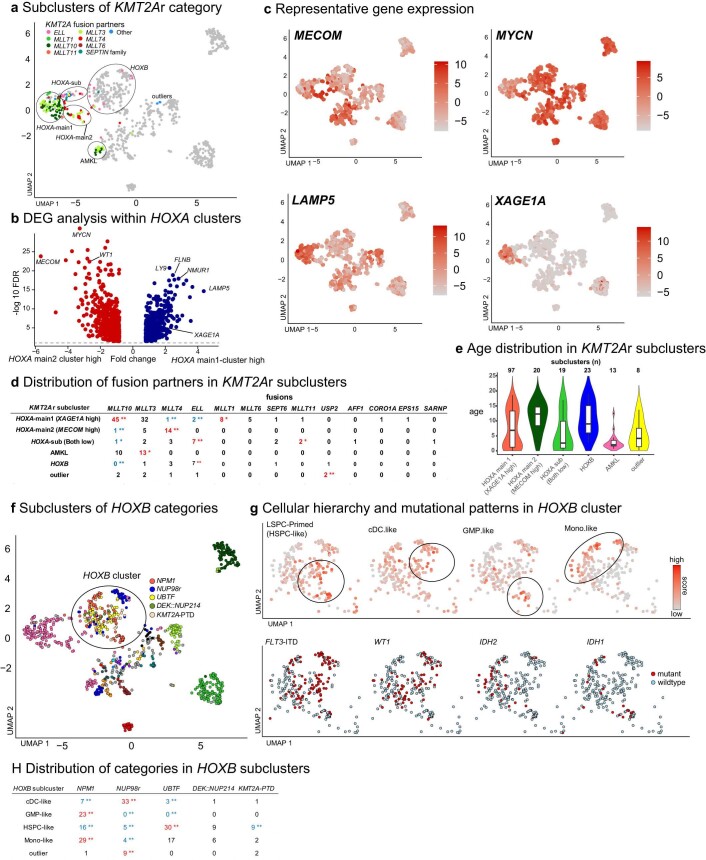
Extended Data Fig. 7. Molecular heterogeneity among HOXA and HOXB groups.
a. UMAP plot showing the distribution of fusion partners of KMT2Ar among different clusters. The dot colors denote fusion partners. b. A volcano plot showing differentially expressed genes (DEG) between the HOXA-main1-2 clusters. Genes with absolute fold change > 2 and FDR < 0.05 are considered DEGs (red: HOXA-main2 cluster high, blue: HOXA-main1 cluster high). Representative gene names are shown. c. Expression of representative DEGs on UMAP plot. The dot colors represent the relative expression of the genes. d. The association of fusion partners of KMT2Ar among different clusters. The statistical significance of the enrichment and exclusivity were assessed by two-sided Fisher’s exact test followed by the Benjamini-Hochberg adjustment (*P < 0.05, **q < 0.05, blue: exclusive, red: enriched). e. Distribution of age at diagnosis among KMT2Ar different clusters. The colors of violin plots represent clusters and lines of the box represent 25% quantile, median, and 75% quantile. The upper whisker represents the higher value of maxima or 1.5 x IQR, and the lower whisker represents the lower value of minima or 1.5 x IQR. Dots represent outliers. f. UMAP plot highlighting molecular categories in the HOXB cluster. The dot colors denote molecular categories. g. Cellular hierarchy scores represented by the color (top) and patterns of frequent mutations (bottom) in the HOXB cluster. Circles in the top highlight clusters with high hierarchy scores. Blue and red dots in the bottom show mutational status. HSPC: hematopoietic stem and progenitor cell. h. The association of molecular categories and HOXB subclusters. The statistical significance of the enrichment and exclusivity were calculated and shown as in d.