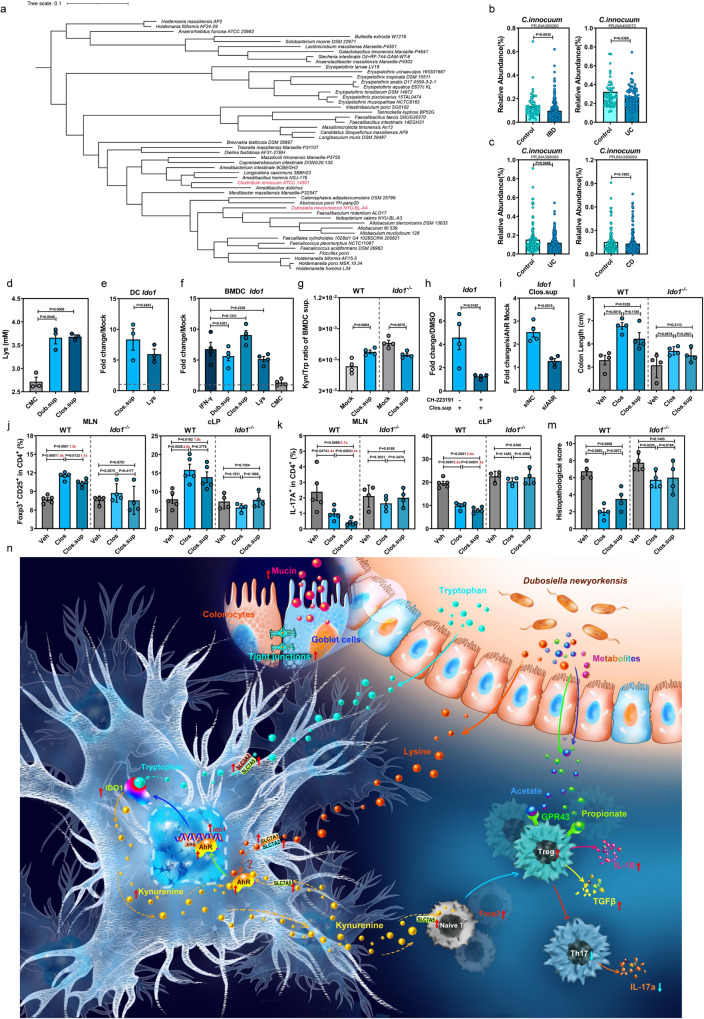
Fig. 7. The human homolog of D. newyorkensis, Clostridium innocuum, generates L-Lys and protects against DSS-induced colitis by activating IDO1 expression in DCs.
a Phylogenic analysis of the Erysipelotrichaceae family and the Clostridium innocuum isolate. b, c Relative abundance of C. innocuum (Clos) in fecal samples from healthy individuals and patients with inflammatory bowel disease (IBD) (b), and specifically ulcerative colitis (UC) and Crohn’s disease (CD) (c). Cohort PRJNA389280 (healthy, n = 55; IBD, n = 221), cohort PRJNA400072 (healthy, n = 55; IBD, n = 73), cohort PRJNA398089 (healthy, n = 178; UC, n = 155; CD, n = 269). d Levels of Lys in the culture supernatants of Dub (Dub.sup) or Clos (Clos.sup) (n = 3). e qPCR of Ido1 mRNA in the Lys- or Clos.sup-treated human DCs extracted from peripheral blood mononuclear cells at 18 h post-treatment (n = 3). f qPCR of Ido1 mRNA at 18 h post-treatment in wild-type C57BL/6J mouse (WT) bone marrow-derived dendritic cells (BMDCs) treated with 8 mM Lys, 20 ng/mL IFN-γ, Dub.sup, Clos.sup or chopped meat carbohydrate broth (CMC) (n = 4). g Kyn/Trp ratio of Clos.sup-treated, or nontreated BMDCs extracted from WT or Ido1−/− mice at 18 h post-treatment (n = 4). h Expression of Ido1 in mouse BMDCs treated with Clos.sup, CH-223191, or a combination of CH-223191 and Clos.sup (n = 4). i Expression of Ido1 in mouse BMDCs transfected with negative control (siNC) or AhR-specific small interfering RNA (siAhR) and treated with Clos.sup at 18 h post-transfection (n = 4). j–m Conventional WT or Ido1−/− mice were colonized with 109 CFU of Clos or treated with Clos.sup and subjected to DSS administration. At D7 post-DSS administration, CD25+Foxp3+Tregs (j) and IL-17+ CD4+ T cells (k) in mesenteric lymph nodes (MLN) and colonic lamina propria (cLP) (WT, n = 5; Ido1-/−, n = 4), colon length (l) and histopathological changes by HE staining (m) were examined at D7 post-DSS treatment (n = 4). (n) Schematic illustration of the D. newyorkensis metabolite Lys protecting against DSS-induced colitis by rebalancing Treg/Th17 response through the activation of the AhR-IDO1-Kyn circuitry. Dashed lines at 1 indicate that the treatments have equal value as normalized controls. Results are representative of data generated in at least two independent experiments and are expressed as mean ± SEM, and 2-sided P-values were examined by the Student’s t-test. Source data are provided as a Source Data file.