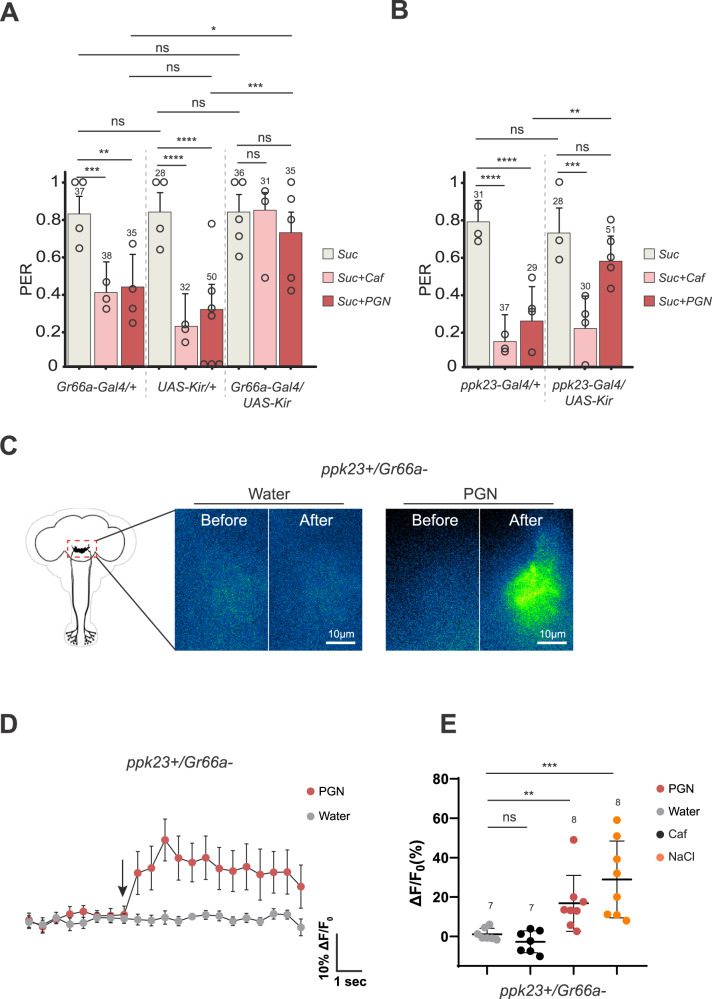
Fig. 2. Adult fly aversion to PGN requires functionally active Gr66a+ and ppk23+ neurons.
Impairing the activity of Gr66a+ (A) or ppk23+ (B) neurons via UAS-Kir2.1 abrogates the aversion to PGN. PER index of flies to control solutions of sucrose 1 mM and sucrose 1 mM + caffeine 10 mM and to sucrose 1 mM + PGN from E. coli K12 at 200 µg/mL. C–E ppk23 + /Gr66a- neurons respond to stimulation with PGN on the labellum. Real-time calcium imaging using the calcium indicator GCaMP6s to reflect the in vivo neuronal activity of ppk23 + /Gr66a- neurons (Gr66aLexA; LexAopGal80; ppk23Gal4/UAS-GCaMP6s) in adult brains of flies whose proboscis has been stimulated with PGN. The expression of LexAop-Gal80 antagonizes the activity of Gal4, thus preventing the expression of GCaMP6s in Gr66a + /ppk23+ neurons. C Representative images showing the GCaMP6s intensity before and after addition of either the control water or the peptidoglycan. D Averaged ± SEM time course of the GCaMP6s intensity variations (ΔF/F0 %) for ppk23 + /Gr66a- neurons. The addition of water (n = 7 flies) or peptidoglycan at 200 µg/mL (n = 8 flies) at a specific time is indicated by the arrow. E Averaged fluorescence intensity of negative peaks ± SEM in response to water (n = 7), caffeine 10 mM (n = 7), Sodium chloride 250 mM (n = 8) or peptidoglycan from E. coli K12 at 200 µg/mL (n = 8). For (A, B), PER index is calculated as the percentage of flies tested that responded with a PER to the stimulation ± 95% CI. The number of tested flies (n) is indicated on top of each bar. For each condition, at least 3 groups with a minimum of 10 flies per group were used. ns indicates p > 0.05, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001 two-sided Fisher Exact Test. In (E) ns indicates p > 0.05, ** indicates p < 0.01, *** indicates p < 0.001, non-parametric t-test, two-tailed Mann-Whitney test. Further details including raw data and exact p-values can be found in the source data file.