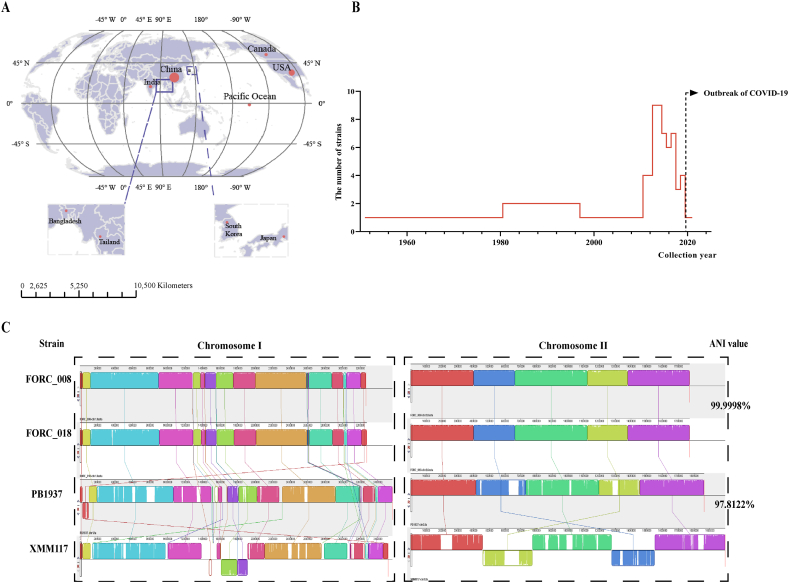
Fig. 1.
The spatiotemporal distribution of V. parahaemolyticus and genome conservation analysis. (A) The main collection regions of V. parahaemolyticus strains. The red circles represent the separation regions of V. parahaemolyticus, and their size represents the number of strains collected in that region. (B) The year range of V. parahaemolyticus isolation. (C) Genetic collinearity analysis of maximum and minimum ANI value genomes. Left: The chromosome Ⅰ structure of the four strains. Right: The chromosome Ⅱ structure of the four strains. Strains FORC_018 and FORC_008 had the highest ANI values, and exhibited the highest similarity in genetic structure (the above two chromosome structure diagrams). The genome structure of the other two strains showed slight changes with XMM117 mainly having two inversions in two chromosomes while PB1937 had translocations in chromosome Ⅰ (the two below ones). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)