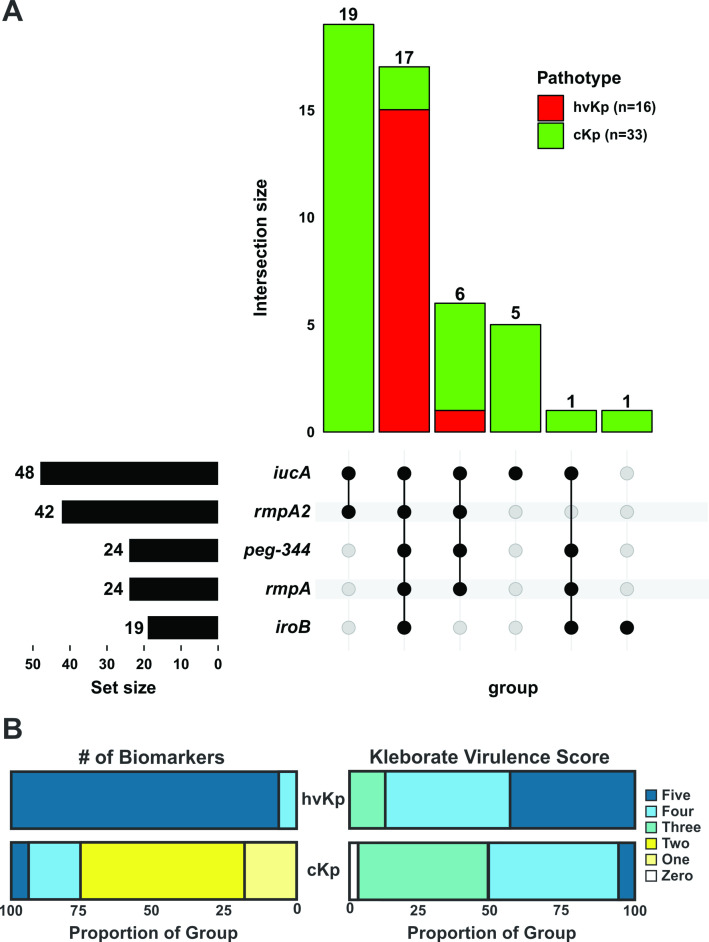
Fig 3.
Biomarker distribution Panel A. UpSet graph for the 49 strains comprising hvKp (n = 16) and cKp (n = 33) was generated to facilitate the visualization of the presence or absence of the biomarkers iucA, iroB, peg-344, rmpA, and rmpA2. Black circles designate the presence of a given marker, whereas gray circles designate its absence. Pathotypes are color coded. The number above each bar represents the number of strains that possess that marker configuration. Panel B: Results are shown as the distribution in proportions of each biomarker count (1–5) and Kleborate virulence score (0–5) for the hvKp and cKp cohorts. The biomarkers count is the presence of some combination of iucA, iroB, peg-344, rmpA, and rmpA2. The Kleborate virulence score is calculated by the presence of the genes that encode aerobactin (three points), yersiniabactin (one point), and colibactin (one point).