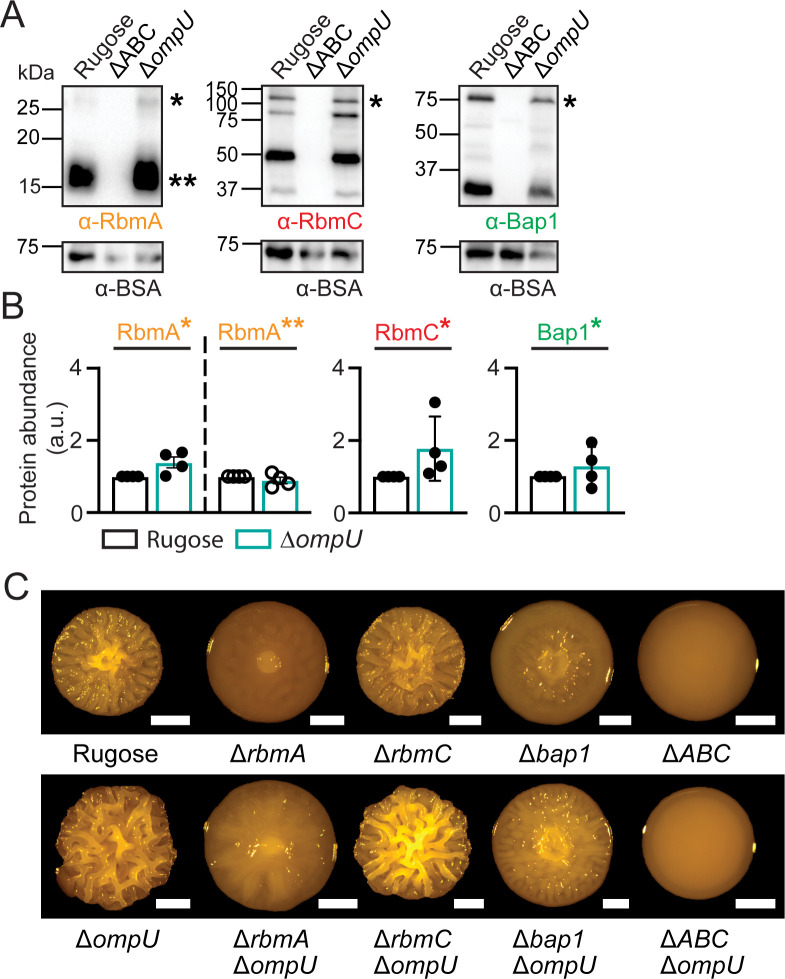
Fig 3.
OmpU does not change matrix protein abundance and affects biofilm colony architecture independently of RbmA, RbmC, and Bap1. (A) Representative immunoblots showing biofilm matrix proteins. Blots were probed against RbmA (left), RbmC (middle), and Bap1 (right) antibodies; single asterisk (*) indicates full-length protein [predicted sizes are 32.5 kDa (RbmA), 114.8 kDa (RbmC), and 82.9 (Bap1)], while double asterisk (**) indicates a proteolytically cleaved variant of RbmA (predicted size 22 kDa). ΔABC stands for the triple rbmA rbmC bap1 deletion strain. Bovine serum albumin (BSA) was added to each sample at equal concentrations, and blots were probed with BSA (predicted size 66.5 kDa) antibodies. Protein ladder sizes in kDa are indicated on the left side of each immunoblot. (B) Analysis of the abundance of the full-length and processed matrix proteins in the biofilm matrix of rugose ΔompU strains. Protein accumulation was analyzed using ImageJ-Fiji software based on at least two biological replicates with two technical replicates per biological replicate. Error bars represent the standard error of the mean. (C) Colony corrugation phenotypes of strains with indicated genotypes after 72 h of growth at 30°C. Images are representative of two biological replicates, with three technical replicates per biological replicate. Scale bars = 1 mm.