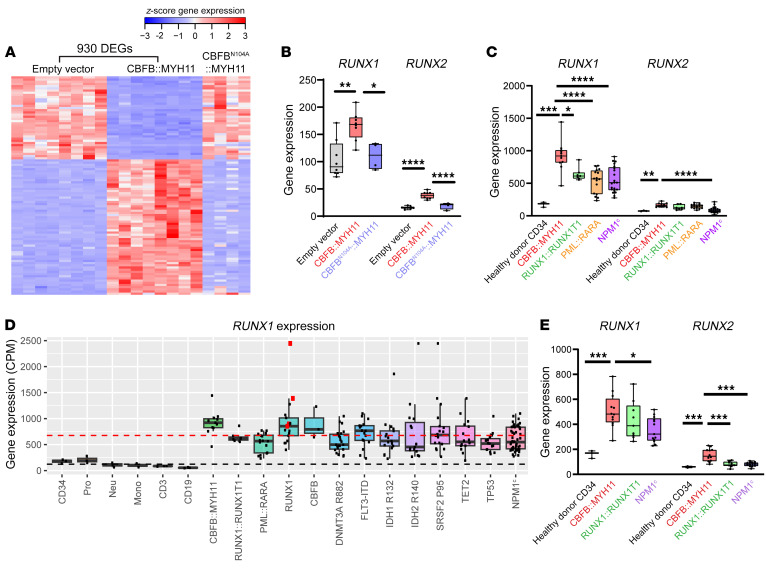
Figure 7. RUNX1 is dysregulated in CBFB::MYH11 AML.
(A) Empty vector (EV; n = 8), CBFB::MYH11 (n = 8), or CBFBN104A::MYH11 (n = 4) was retrovirally expressed in murine hematopoietic cells, and RNA-Seq was performed at days 4 and 7. A heatmap of the top 100 DEGs shows that the CBFB::MYH11 transcriptional signature is abrogated by the CBFBN104A mutation. (B) Runx1 and Runx2 are upregulated in CBFB::MYH11 (red) but not CBFBN104A::MYH11 (blue) cells relative to EV cells (gray), suggesting that upregulation may be due to a CBF-sensing feedback loop. One-way ANOVA between all samples, *P < 0.05, **P < 0.01, ****P < 0.0001, nonsignificant comparisons unlabeled. Each point represents an individual sample, bar indicates mean, box indicates 95% confidence interval, whiskers indicate value range. (C) Human TCGA AML RUNX1/2 RNA-Seq data for healthy donor CD34+ (black, n = 3) and CBFB::MYH11 (red, n = 11), RUNX1::RUNX1T1 (green, n = 7), PML::RARA (yellow, n = 16), or NPM1c-mutant AMLs (purple, n = 21). RUNX1 was overexpressed in CBFB::MYH11 samples relative to all other subgroups; RUNX2 was overexpressed relative to CD34+ and NPM1c subgroups. One-way ANOVA between all subgroups, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, nonsignificant/non-CBFB::MYH11 comparisons not shown. (D) Human TCGA AML RUNX1 RNA-Seq data for healthy donor (CD34+; Pro, promyelocytes; Neu, neutrophils; Mono, monocytes; CD3, T cells; CD19, B cells) or AML samples with the indicated oncofusion/mutation. Red squares, biallelic RUNX1 mutations; black dashed line, mean healthy donor cell RUNX1 expression; red dashed line, mean AML RUNX1 expression. All AML samples have upregulated RUNX1 relative to healthy donor cells; AML samples with CBFB::MYH11, RUNX1, or CBFB mutations have mean RUNX1 expression above the AML average. (E) RNA-Seq data for RUNX1/2 from an independent cohort of healthy donor CD34+ (black, n = 3) and CBFB::MYH11 (n = 12, red), RUNX1::RUNX1T1 (n = 9, green), and NPM1c AMLs (n = 13, purple), confirming higher levels of RUNX1/2 expression in CBFB::MYH11 AMLs. One-way ANOVA between all subgroups, *P < 0.05, ***P < 0.001, nonsignificant/non-CBFB::MYH11 comparisons not shown.