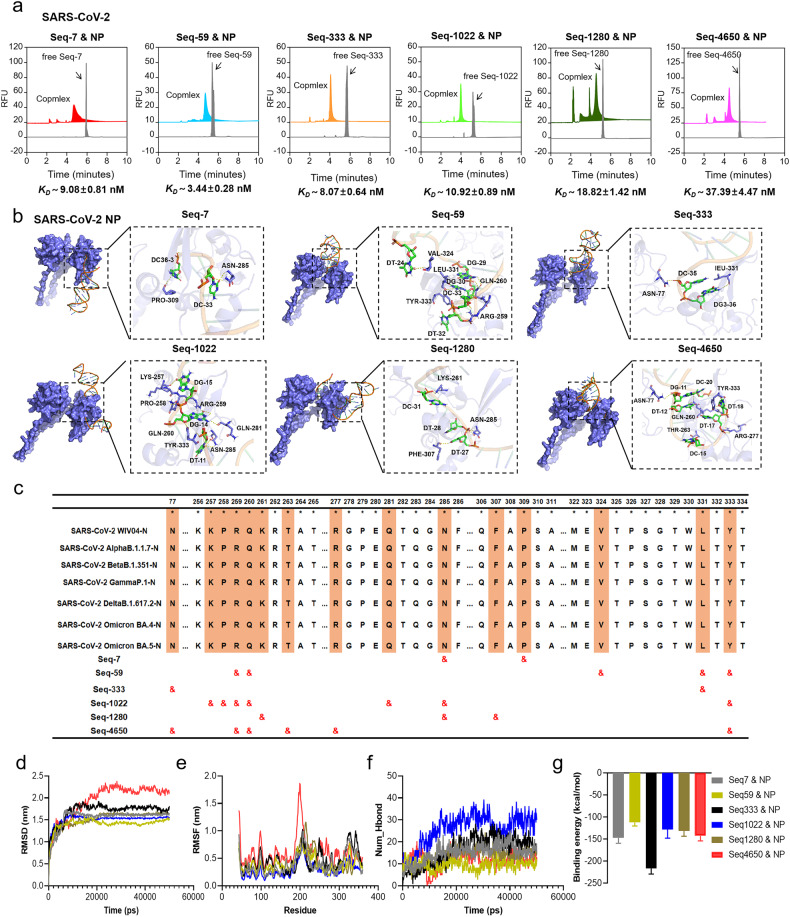
Fig. 2.
Characterization of the affinity and the Binding sites prediction between aptamers and N protein of SARS-CoV-2. a Affinity and specificity characterizations between aptamers and N protein of SARS-CoV-2 were performed via CE, n = 3 per group. b The results of molecular docking to predict and simulate the binding sites of aptamers to SARS-CoV-2 NP using Discovery Studio software. The NP is shown with residue names and identifier. c The binding sites of six ssDNA aptamers to NP in different SARS-CoV-2 mutants. d–g Molecular dynamics simulation results between aptamers and NP of SARS-CoV-2. d Root mean square deviation (RMSD). e Root mean square fluctuation (RMSF). f Number of intermolecular hydrogen bonding (H-bonds). g The binding energy of aptamers and NP of SARS-CoV-2