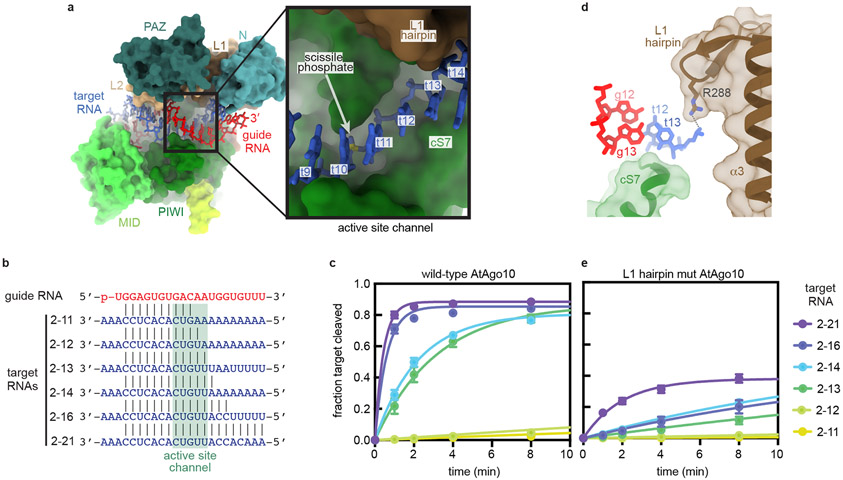
Fig. 4. Structural basis for RNA Slicing.
a. Surface representation of the AtAgo10 central-duplex model. Inset shows t9–t13 (blue sticks) docked in the active site channel (guide RNA omitted for clarity). b. Base pairing schematic of guide and target RNAs used in slicing reactions in panels c and e. Target nucleotides falling within the active site channel are indicated. c. Fraction of target RNAs cleaved by a saturating excess of wild-type AtAgo10-guide complex versus time. d. Side-view of the end of the active site channel where the L1 hairpin secures t13 against cS7. e. Fraction of target RNAs cleaved by a saturating excess L1-hairpin-mut AtAgo10-guide complex versus time. Data points are the mean values of n=3 independent experiments. Error bars indicate SEM.