Fig. EV2. Reconstruction of the AtAgo10-guide complex.
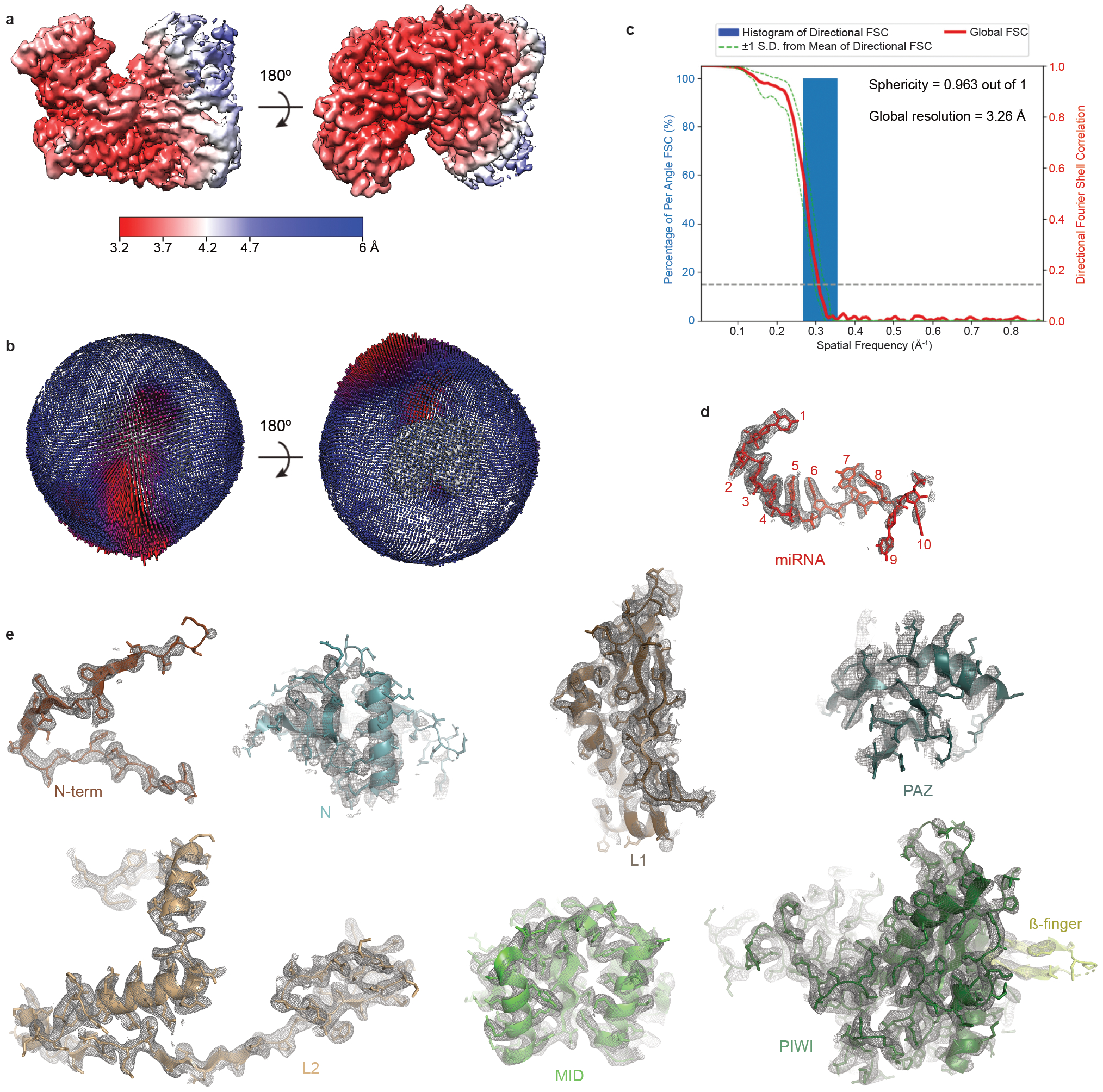
a. The final 3D map for the AtAgo10-guide RNA complex colored by local resolution values, where the majority of the map was resolved between 3.3 Å and 3.7 Å, and the map covering the more mobile PAZ and N domains has lower resolution. b. Angular distribution plot showing the Euler angle distribution of the AtAgo10-guide particles in the final reconstruction. The position of each cylinder corresponds to the 3D angular assignments and their height and color (blue to red) correspond to the number of particles in that angular orientation. c. Directional Fourier Shell Correlation (FSC) plot representing 3D resolution anisotropy in the reconstructed map, with the red line showing the global FSC, green dashed lines correspond to ±1 standard deviation from mean of directional resolutions, and the blue histograms correspond to percentage of directional resolution over the 3D FSC. d. EM density (mesh) quality of the guide RNA (sticks). e. Individual domains of AtAgo10 fit into the EM density, EM density shown in mesh; molecular models (colored as in Fig. 1) shown in cartoon representation with side chains shown as sticks.
