Abstract
Investigations of the bacterial family Micromonosporaceae have enabled the development of secondary metabolites critical to human health. Historical investigation of bacterial families for natural product discovery has focused on terrestrial strains, where time-consuming isolation processes often lead to the rediscovery of known compounds. To investigate the secondary metabolite potential of marine-derived Micromonosporaceae, 38 strains were sequenced, assembled and analysed using antiSMASH and BiG-SLiCE. BiG-SLiCE contains a near-comprehensive dataset of approximately 1.2 million publicly available biosynthetic gene clusters from primarily terrestrial strains. Our marine-derived Micromonosporaceae were directly compared to BiG-SLiCE’s preprocessed database using BiG-SLiCE’s query mode; genetic diversity within our strains was uncovered using BiG-SCAPE and metric multidimensional scaling analysis. Our analysis of marine-derived Micromonosporaceae emphasizes the clear need for broader genomic investigations of marine strains to fully realize their potential as sources of new natural products.
Keywords: biosynthetic gene cluster, drug discovery, genomics, marine-derived, natural products
Abbreviations
AAI, average amino acid identity; ANI, average nucleotide identity; BGC, biosynthetic gene cluster; GCF, gene cluster family; MDR, multi-drug-resistant; MDS, multidimensional scaling; NP, natural product; NRPS, non-ribosomal peptide synthetase; PKS, polyketide synthase; RiPPs, ribosomally synthesized and post-translationally modified peptides; SSN, sequence similarity network.
Impact Statement
The increasingly deadly threat to humanity posed by drug-resistant and completely new infectious diseases continues to drive efforts to expedite and expand efforts to identify new antimicrobial agents as therapeutics and drug leads. Microbial producers of, as yet, undiscovered new small molecules with medicinal potential have originated historically from terrestrial sources. However, it is becoming increasingly apparent that marine environments harbour repositories of microbial small molecule producers that rival, if not surpass, the structural and biosynthetic diversities of terrestrial organisms. Our work here supports this notion, and given recent advances in bioinformatics, culturing methodologies and available analytical instrumentations, seeks to illuminate the potential of marine-derived microbes as beacons for discovery of new anti-infective drug leads. Careful comparisons of marine genomics information to databases obtained from terrestrial microbes bears out the clear need to better exploit marine-derived microbes as points of discovery and inspiration. This work seeks to impact the field of microbial genomics and drug discovery by showcasing marine-derived microbes as high-priority opportunities for the discovery of new and probably novel biosynthetic machineries and their resulting natural products.
Data Summary
Large datasets (mostly as tables) and special files can be found in Zenodo (https://zenodo.org/) and Figshare (https://figshare.com/) under the following links: https://zenodo.org/record/8137859 and https://doi.org/10.6084/m9.figshare.24492253.v1. In particular, the following collections of data for this paper are included:
Data S1: A folder with all the fasta files, representing the 42 strains (41 Micromonosporaceae, 1 Streptomycetaceae).
Data S2: A folder with all the .gbk files for the biosynthetic gene cluster (BGC) regions predicted by antiSMASH v5.1.1. These files were used as inputs for BiG-SCAPE and BiG-SLiCE.
Data S3: A folder with all the .gbk files for the BGC regions predicted by antiSMASH v6.1.0.
Data S4: A folder containing all the Quast outputs for the 42 strains.
Data S5: A folder containing all the BUSCO outputs for the 42 strains. Example scripts are provided for scraping relevant information from the individual BUSCO outputs.
Data S6: A folder containing GTDB (Genome Taxonomy Database) classification results, and species-level grouping results using FastANI (95 % cutoff).
Data S7: A folder containing an Interactive Tree of Life (iTOL)-compatible bar chart annotation using antiSMASH v5.1.1 BGC region information.
Data S8: A folder containing a Word document that describes the parameters used with Ubuntu WSL (Windows Subsystem for Linux) on the command line for programs antiSMASH v6.1.0, BiG-SCAPE v1.1.2 and BiG-SLiCE v1.1.1. Also included are parameters for metric MDS in python. An example script is also provided for batch queries of BGCs against BiG-SLiCE v1.1.1’s pre-processed dataset of ~1.2 million BGCs.
Data S9: A folder containing the BiG-SCAPE visualization of the 38 Micromonosporaceae (post-QC filtering, excluding WMMA1363, WMMB482, WMMB486 and WMMC500) in Cytoscape.
-
Data S10: A folder containing:
The pre-processed dataset of 1.2 million BGCs from BiG-SLiCE.
All report folders generated by BiG-SLiCE for the 779 Micromonosporaceae BGCs queried against the 1.2 million BGCs.
The results data.db and associated folders for the pre-processed dataset of 1.2 million BGCs.
Data S11: A folder containing the scripts necessary to regenerate the figures and perform independent analyses, and the relevant data used for the analyses.
Data S12: A folder containing the NCBI blast query used to compare WMMD1947 region 12 against WMMD1120 region 14 using antiSMASH v6.1.0.
Data S13: A folder containing the files pertaining to RiPP subclasses, BiG-SLiCE BGCs annotated with BiG-SCAPE information, and MIBiG BGCs identified as being related to BGCs in our dataset.
In addition to having provided these materials through Zenodo and FigShare, the authors confirm that all supporting data, code and protocols have been provided within the article or through supplementary data files.
Introduction
Drug-resistant infectious diseases have been recognized for decades as a growing threat to humanity [1, 2]. More recent crises such as sudden respiratory syndrome (SARS), monkey pox and, most dramatically, the coronavirus disease 2019 (COVID-19) pandemic have exacerbated the issue significantly due to the increased use of antimicrobials and the predictable acceleration of healthcare-associated drug-resistant infections in US hospitals [3, 4]. In tandem with these realizations, it has also long been recognized that bacterially derived secondary metabolites (natural products, NPs) constitute an idealized repository of new drug leads with the potential to display novel mechanisms of action, and thus the ability to circumvent current drug resistance mechanisms in pathogens [2, 5, 6]. However, decades of mining terrestrial sources for useful NPs have reduced the likelihood of identifying truly new and novel NPs, despite truly remarkable technical advances that have streamlined drug discovery processes [5, 7]. In contrast, recent efforts have revealed marine-derived microbes to be extremely attractive and, now, tractable sources; this is especially true for actinobacterial populations [7–13]. For instance, the marine actinomycete genus Salinispora was prioritized for novel NP explorations and subsequently enabled the discovery of lomaiviticins, salinosporamides and other antibacterial compounds [13].
Despite their inability to grow in deionized water compared to seawater, Salinispora have been disproportionately studied within the marine-derived NP community; the breadth of Salinispora studies to date has no doubt been a result of their well-noted and chronologically early recognition as sources of NP originality [13–16]. Perceived limitations of Salinispora, as a genus, and technological advances of the last 10 years, have inspired recent campaigns to aggressively evaluate other marine-derived bacteria as sources for new NP scaffolds [12].
For example, phylogenetic analyses of brackish water sponges unveiled proportionally more Micromonospora spp. compared to Streptomyces spp., contrasting earlier analyses of soil environment isolates [9, 17]. Moreover, comparisons of brackish water habitats and tropical reef-type habitats have revealed an abundance of four specific genera: Streptomyces spp., Micromonospora spp., Solwaraspora spp. and Verrucosispora spp. [9]. With distributions of bacterial taxa differing across environments, notable metabolic diversity within Micromonospora spp., and the historical emphasis on Streptomyces spp. exploration, it is now clear that investigations of marine Micromonosporaceae are likely to unveil NPs that differ from those of terrestrial strains, including both general terrestrial bacterial strains and terrestrial Micromonosporaceae Micromonospora spp. [9, 17, 18]. (This study showed that members of the family Micromonosporaceae were more abundant among cultivated bacteria from tropical ecosystems versus brackish ones. In addition, metabolomics initiatives have shown their metabolites to differ substantially from those of brackish Streptomyces.)
Notably, the potential of Micromonospora to generate antimicrobials was first recognized in 1947 with the discovery of the polyketide micromonosporin and subsequently underscored with the discovery of gentamicin (Gentocin, Garamycin) from Micromonospora purpurea in 1963; importantly, gentamicin is listed by the World Health Organization (WHO) as an essential and critically important medication [19–21]. In the time intervening gentamicin’s discovery and now, over 740 antibiotics have been discovered from Micromonospora strains although remarkably few have received clinical approval. Not surprisingly, the metabolic capacities of Micromonospora family members and their impact on drug discovery initiatives have been the subject of several outstanding reviews in recent years [7, 18, 22]. Gentamicin, netilimicin, plazomicin (Zemdri), isepamicin, neomycin (neo-Fradin, neo-Tab) and sisomicin (bactoCeaze, Ensamycin) represent, by far, the most clinically successful Micromonospora-derived agents and all remain in service today to differing extents based on geographical location and specific antimicrobial applications [18, 22]. In addition to aminoglycosides, Micromonospora are known to produce antimicrobials bearing macrolide, ansamycin, everninomicin and actinomycin scaffolds [7, 18, 22, 23]. In addition to serving as sources of antimicrobials with activity against ‘susceptible pathogens’, Micromonospora have also yielded metabolites with potent activity against multiple-drug-resistant (MDR) microbes. For instance, turbinmicin, reported in 2020 from Micromonospora WMMC-415, displays broad-spectrum activity against MDR fungal pathogens, notably Candida auris and Aspergillus fumigatus [24, 25] and is currently in clinical development. In addition, Micromonospora have been shown to produce the enediyne-based DNA-damaging agents calicheamicin, yangpumicins and dynemicin; all hail from Micromonospora and have been applied to the creation and use of human therapeutics to differing extents [7, 18, 22, 26–28]. Thus, it is now abundantly clear that Micromonospora and associated family members of the Micromonosporaceae represent outstanding resources for NP discovery, and highlight environments that produce these bacteria as primary investigative targets.
Concurrent with shifting views on the importance of marine vs. terrestrial NP repositories has been the continuous advancement of technologies aimed at acquiring and exploiting genetic information, specifically the development of high-throughput DNA sequencing methods. Compared to traditional 16S rRNA gene sequencing and phylogenetic strain classification, whole genome sequencing as used by the Genome Taxonomy Database has allowed for enhanced disambiguation of genomes/organisms [29, 30]. Average nucleotide identity (ANI), accessible via whole genome sequencing, now enables novel bacteria to be better understood relative to publicly available literature; one result of this advance has been the ability to delineate classically understood genera into more representative genera [30]. Additional genomic information acquired via whole genome sequencing has shed significant light on orphan biosynthetic gene clusters (BGCs) housed within bacteria and fungi [31]. BGCs represent groupings of genes involved with the production of secondary metabolites, thus linking genomics to the production and intracellular use of bioactive NPs [32]. Relatedly, the refinement of antiSMASH (antibiotics and Secondary Metabolite Analysis Shell) has enabled the expanded exploitation of bacterial genomes in the search for antimicrobial NPs [33]. In conjunction, the construction of databases from public literature sources such as MIBiG (Minimum Information about a Biosynthetic Gene cluster) has allowed for improved use of already described BGCs in the search for new NPs [32].
Importantly, tools such as antiSMASH and MIBiG can be utilized by downstream software such as BiG-SCAPE (Biosynthetic Gene Similarity Clustering and Prospecting Engine) and BiG-SLiCE (Biosynthetic Gene clusters – Super Linear Clustering Engine) to create sequence similarity networks (SSNs) across strain collections [32–35]. These genome-guided computational tools enable one to identify uncharacterized BGCs and their related products across large collections of strains and employ both publicly available literature and privately held datasets [36]. Such an approach minimizes the likelihood of NP rediscovery, a well-known hurdle to effective drug discovery efforts and maximizes currently known correlations of genomic information to small molecule structure and function. These newfound capabilities are ideally suited to performing critical analyses regarding new NP potentials of any collection of microbial NP producers one might wish to interrogate.
To date, a broad analysis of marine Micromonosporaceae genomes, BGCs related to the production of NPs and their relation to public databases has been lacking. Hence, we provide here a comparative genomic analysis of marine Micromonosporaceae with the intention of identifying truly novel BGCs. Our analyses revealed the biosynthetic diversity of marine Micromonosporaceae, especially in comparison to publicly curated databases. Interestingly, we identified four new species belonging to Micromonospora_E, which is a new genus within the GTDB-tk2 database. These species represent a 4-fold increase in known Micromonosporaceae Micromonospora_E compared to the 311 480 bacterial strains stored in GTDB-tk2 from publicly available data. Further investigations necessitated the development of a novel methodology to visually explore BGC similarity across an individual strain collection and against publicly available data, to prioritize unique BGCs for identification and investigation . This study showcases: (a) the utility of prioritizing BGCs based on novelty, relative to large existing databases; and (b) the potential of renewed investigations into marine Micromonosporaceae as an outstanding repository for NP scaffolds.
Methods
Strain isolation and extraction
Bacterial strains were isolated using previously published methodologies [37]. 16S rRNA gene sequences were extracted using standard methods [37]. DNA sequences were extracted using standard protocols for our laboratory [38].
Strain sequencing, assembly and validation
Next-generation sequencing was performed with PacBio Sequel platforms using two Sequel single-molecule real-time (SMRT) cells [University of Wisconsin, Madison (UW-Madison), Biotechnology Center]. PacBio data were corrected, trimmed and assembled with Canu v1.8 [39] using the parameter ‘genomeSize=8m’ (UW-Madison, Center for High Throughput Computing). BUSCO v5.4.3 [40] and QUAST v5.0.2 [41] were used to assess each genome assembly based on completeness and quality respectively, and the results are listed in Data S4 and S6 (available in the online version of this article).
Annotation of biosynthetic gene clusters
To identify BGCs related to secondary metabolism, genome sequences in Fasta format were annotated by installations of antiSMASH v5.1.1 [42] and antiSMASH v6.1.0 [43]. For antiSMASH v5.1.1, a minimal run omitting additional features was carried out using Antibiotic Resistant Target Seeker Version 2 (ARTS) [44, 45]. For antiSMASH v6.1.0, BGC prediction was performed with detection strictness relaxed, and extra features KnownClusterBlast, ActiveSiteFinder, SubClusterBlast and RREFinder were active. The full parameters for the antiSMASH runs are listed in Data S8. Detailed information for the 39 strains is listed in Data S4–S6.
A pie plot of overall BGC counts per predicted product type was constructed using seaborn (v0.11.2) [46] in Python (v3.7.12) [47].
Phylogenomics, average identity estimation between genomic pairs and taxonomic classification
We identified 922 single-copy core orthologues between the genomes of 39 isolates using OrthoFinder v2.5.4 [48]. Multiple sequence alignment of protein sequences was performed for each orthologue using muscle v5 [49] and filtered for sites which featured gaps in more than 10 % of samples using trimAI v1.4 [50]. Filtered protein alignments were concatenated to produce an alignment of length 292 788 aa with coordinates for individual alignments of the 922 orthologues used to generate input for IQ-Tree v2.2.0.3 [51] phylogeny reconstruction using an edge-proportional partition model with ModelFinder Plus [52] and visualized using iTOL v6.6 [53] (Fig. 1a). CompareM v0.1.2 (https://github.com/dparks1134/CompareM) was used to assess average amino acid identity (AAI) between the 39 genomes. The Genome Taxonomy Database (GTDB-tk v2) [30] was used to classify taxonomies for the 39 genomes with GTDB release R207 [54], and the taxonomic identification results are listed in Data S6. FastANI [55] was used to group species across the 39 genomes using ANI, and the species grouping results are listed in Data S6.
Fig. 1.
Overall representation of phylogeny and BGCs of Micromonosporaceae type strains included in this work. (a) Phylogenetic tree of 38 Micromonosporaceae strains and one Streptomycetaceae strain (WMMC500). The bar plot represents the antiSMASH v5.1.1 identified BGCs, and colours represent the predicted BGC product type. Bar, 1 substitution per position. (b) Pie chart describing the 779 BGCs identified in the 38 Micromonosporaceae strains according to antiSMASH v5.1.1’s predicted product type.
Analysis of intra-BGC diversity using marine Micromonosporaceae
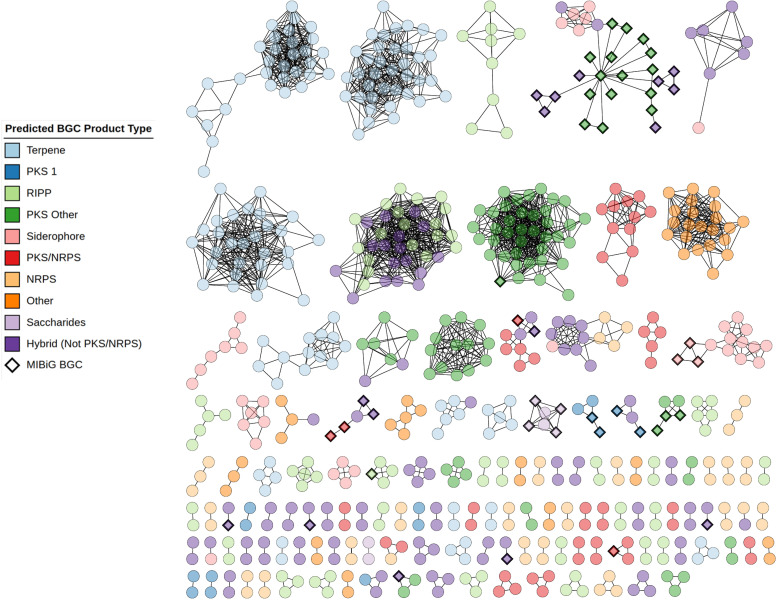
Utilizing the Micromonosporaceae antiSMASH v5.1.1 GenBank files, pairwise distance across predicted BGCs and SSNs was constructed using BiG-SCAPE tool v1.1.0 [34]. This run of BiG-SCAPE was performed at a default cutoff of 0.3 with additional features such as: including BGCs from the MIBiG database v2.1 [56], constructing an all-vs-all distance network mixing all BGC product classes, including BGCs that do not have a distance lower than the cutoff distance specified, and including BGCs with hybrid predicted products into each subclass network. The full parameters for the BiG-SCAPE analysis are listed in Data S8. The network output of BiG-SCAPE was imported into Cytoscape v.3.9.1 [57] (Fig. 2). The BGC product annotations generated by antiSMASH v5.1.1 were categorized based on BiG-SCAPE cluster classes and were added to the Cytoscape visualization. The MIBiG BGC product types were identified using minimal runs of antiSMASH v5.1.1 through ARTS [44, 45], manually annotated using BiG-SCAPE cluster classes categories, and added as annotations to the Cytoscape network.
Fig. 2.
Sequence similarity network (SSN) produced by BiG-SCAPE when analysing Micromonosporaceae strains, visualized and annotated with CytoSCAPE. Nodes represent individual BGCs. BGC types are coloured according to the colour key. Micromonosporaceae BGCs are represented as circles and MIBiG BGCs are represented as diamonds. Singletons (196 BGCs) have been removed from the visualization, and are shown in Fig. S181.
Analysis of BGC diversity against terrestrial bacteria
Micromonosporaceae antiSMASH v5.1.1 outputs were queried against the pre-processed dataset of 29 955 gene cluster family (GCF) models in BiG-SLiCE v1.1.1 [35] using a clustering threshold value of 900. The 29 955 GCF models were previously computed by Kautsar et al. [35] using a clustering threshold distance of 900 to group 1 225 071 BGCs into GCFs. For each BGC–GCF pairing, a membership score (distance) was generated, resulting in 779 BGCs having an associated 29 955 distances for each possible GCF. These distances corresponding to BGC–GCF instances were ranked from lowest to highest to generate the top-X hits, where X represents the X-best GCF hits for each BGC, with a lower distance indicating higher confidence in similarity. The full parameters for the BiG-SLiCE runs are listed in Data S8. The file-based SQL database was accessed in Python using the SQLite3 library. The best-performing GCF for each Micromonosporaceae BGC was investigated for the presence of MIBiG BGCs and the distance values associated with the BGCs and their topmost GCF pair was plotted in Fig. 3.
Fig. 3.
Scatter plot of Micromonosporaceae and Streptomycetaceae BGCs queried against BiG-SLiCE (threshold=900). Nodes represent individual BGCs. The horizontal red line indicates the threshold for successful clustering of a BGC into a GCF from BiG-SLiCE. The dots are coloured: (a) orange, if the GCF most similar to that BGC contained a BGC from MIBiG; (b) blue, if the GCF most similar to that BGC did not contain a BGC from MIBiG; and (3) green, if the BGC’s distance to the closest GCF fell above the clustering threshold of 900.
Proposed methodology for identification of novel BGCs
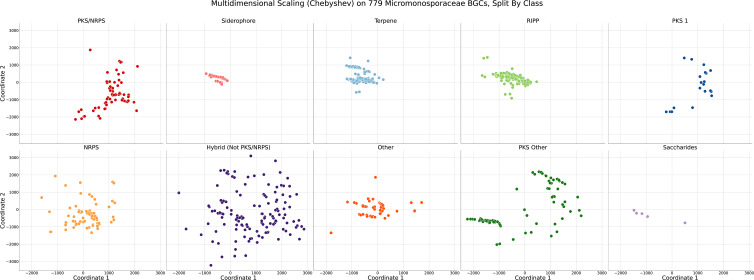
The table of membership scores (distances) between 779 Micromonosporaceae BGCs and the 29 955 GCFs in BiG-SLiCE was transformed into a pairwise distance matrix using the Chebyshev distance metric, resulting in a 779×779 matrix describing the distance between BGCs in relation to the distribution of membership scores (distances) across the GCFs. Metric multidimensional scaling (MDS) was performed as a dimensionality reduction technique to reduce the 779×779 matrix into a two-dimensional space [58]. The metric MDS parameters are described in Data S8, and the corresponding metric MDS plots are visualized in Figs 4 and S1–S10. The MDS scatter plot marker size and hue were scaled based on the average distance of the BGC to the top three GCFs (Figs 5 and S11–S20).
Fig. 4.
Scatter plots of Micromonosporaceae BGCs analysed via multidimensional scaling using Chebyshev pairwise distance. Each dot represents an individual BGC. Distance between BGCs is associated with GCFs in BiG-SLiCE to which they were most similar. Visualization was separated according to predicted BGC product type. The colour code represents the predicted BGC product type.
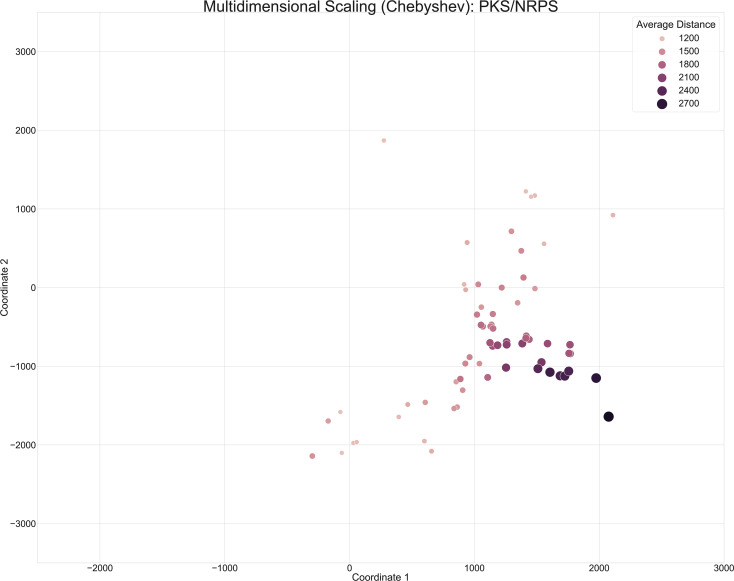
Fig. 5.
Annotated version of the scatter plot of PKS/NRPS Micromonosporaceae BGCs from Fig. 4. The hue and size of the dots were scaled based on the average distance of the BGCs, across the entire dataset, to the top three GCFs in BiG-SLiCE (T=900).
The BGC-GCF table of 779 BGCs and their membership scores (distances) to the 29 955 GCFs in BiG-SLiCE was split into sub-datasets. These sub-datasets were based on the antiSMASH predicted product type of the BGC, resulting in 10 sub-datasets. Each sub-dataset was transformed into pairwise distance matrices, using eight different distance metrics. These metrics were Euclidean, Cosine, Cityblock, Chebyshev, L2, Braycurtis, Canberra and Correlation pairwise distances. The pairwise distance matrices underwent metric MDS to compare our BGCs against each other (Figs 6 and S21–S100). The size and hue of the markers for each BGC in the metric MDS visualizations were scaled based on the average distance of the BGC to the top three GCFs (Figs 7 and S101–S180). The metric MDS parameters are described in Data S8.
Fig. 6.
Improved scatter plots of Micromonosporaceae BGCs, separated by predicted BGC product type prior to analysis via multidimensional scaling with Chebyshev pairwise distance. Distances between BGCs of a specific product type were associated with most similar GCFs in BiG-SLiCE. Predicted BGC product types are colour coded.
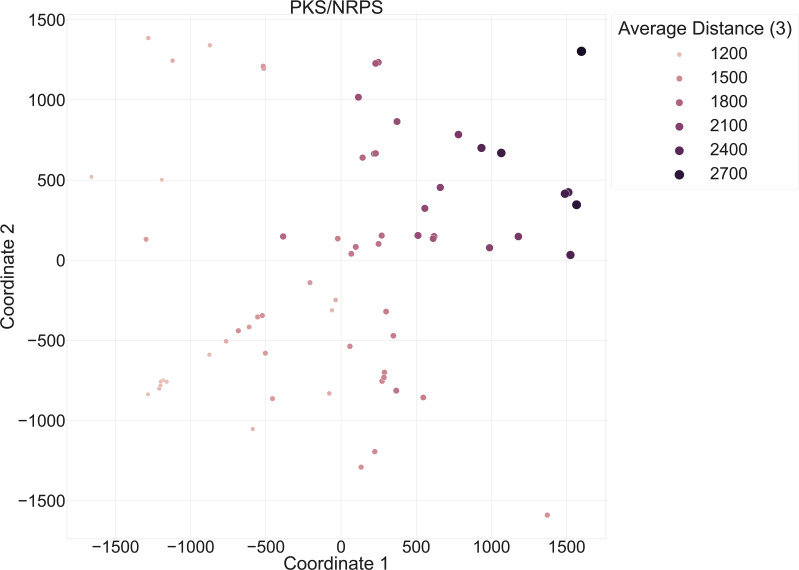
Fig. 7.
Annotated version of the improved scatter plot of PKS/NRPS Micromonosporaceae BGCs from Fig. 6. The hue and size of the dots were scaled based on the average distance of the BGCs, across the predicted BGC product type PKS/NRPS, to the top-3 GCFs in BiG-SLiCE (T=900).
Statistical analysis of BGC similarity distribution across genera
The 570 Micromonospora BGCs and 147 Micromonospora_E BGCs were queried against BiG-SLiCE for the best GCF membership score. A comparison of the distributions of BGCs across genera was constructed using a box plot (Fig. 8; Table S1). The Mann–Whitney U-test was applied to evaluate the statistically significant differences across the genera-specific distributions (Table S1). All analyses utilized a significance level of P<0.05.
Fig. 8.
Box and whisper plots comparing the distribution of distances between individual BGCs and the associated closest GCFs in BiG-SLiCE. The distributions have been separated by genera, with six Micromonospora_E strains and 29 Micromonospora. The red dot represents the means of the distributions. The Mann–Whitney U-test (double-sided) was used to confirm differences in the distributions.
Results and discussion
Novel NPs from primarily terrestrial bacteria have served as the first and last lines of defence against antibiotic-resistant pathogens for over 90 years [2]. However, trend analyses regarding decades of NP research indicate that terrestrial NP repositories are reaching critical exhaustion levels; in particular, the likelihood of discovering bioactive NPs with clinically employable novel mechanisms of action is becoming diminishingly small. Accordingly, drug discovery initiatives of the last 5–10 years are now aggressively pursuing alternative sources of microbially derived NPs with clinical potential. Inspired by our own interest in marine-derived microbes and the increasingly dire rise of antibiotic resistance, we evaluated the family Micromonosporaceae, specifically marine-associated Micromonosporaceae, as a promising source of NPs with bioactivity against clinically relevant pathogens. The approach taken, one that critically evaluates BGC content of assorted pools of microbes, enabled us to determine if marine-associated bacteria are sufficiently distinct from previously characterized terrestrial bacteria to warrant in depth marine-focused metabolomics. In this work, we incorporated genomics-based analytics to uncover potential bioactive secondary metabolites unrelated to known molecules based on comparative analyses of microbial BGC content. However, while the dataset used for this study contains a significant number of Micromonosporaceae sequenced by our research group, it was not explicitly designed to be a diverse set of bacteria based on phylogenetic grounds and does not represent the entire marine-associated family with regard to metabolic potential.
Marine bacterial strain genome characteristics
The genome sequences of the 38 selected Micromonosporaceae strains, one selected Streptomycetaceae strain, associated QUAST (v5.0.2) annotations and their respective BUSCO information are presented in Data S1. The G+C content of the Micromonosporaceae strains varied from approximately 70.41 to 73.94 %. Genome sizes ranged from 6 311 079 to 8 920 919 bp, completeness scores ranged from 98.3 to 100 %, and the number of genes ranged from 5688 to 8739.
Phylogenetic classification of marine bacterial strains
The phylogenetic classifications of the 38 selected Micromonosporaceae strains and one Streptomycetaceae strain were performed using GTDB-tk v2 (Data S6). AAI and ANI analyses were performed to identify shared genera and shared species, with a minimum threshold of 67.66 % AAI being necessary to belong to the same genera, and an upper bound of 95 % ANI to be considered a novel species strain (Data S6). WMMC500 (Streptomycetaceae Streptomyces) was found to be a novel species of Streptomyces. WMMD1047 (Micromonosporaceae) was uncovered as a novel Micromonosporaceae sp. through GTDB-tk v2 taxonomic classification. Utilizing GTDB-tk v2 in conjunction with ANI analysis revealed 18 bacterial strains as belonging to known species of Micromonosporaceae Micromonospora. Continued analysis identified 11 bacterial strains as eight novel species of Micromonosporaceae Micromonospora. Further investigation discovered six bacterial strains belonging to the genus Micromonospora_E, with four novel species of Micromonosporaceae Micromonospora_E identified. The designation Micromonospora_E comes from GTDB-tk2 splitting the current genus of Micromonospora into Micromonospora, Micromonospora_E, Micromonospora_G and Micromonospora_H based on AAI [54]. GTDB-tk2 also revealed the presence of two additional novel species, WMMD1127 (Micromonosporaceae Asanoa, unknown sp.) and WMMD1102 (Micromonosporaceae Plantactinospora, unknown sp.).
Phylogenetic analysis of our 38 selected Micromonosporaceae strains and one Streptomycetaceae strain broadly conformed to expectation. WMMC500 (Streptomycetaceae Streptomyces, unknown sp.) correctly self-identified as a unique clade in our phylogenetic tree and was predicted as a new species of Streptomyces that is distinct from the genomes in GTDB-tk2. This result was expected, as Streptomycetaceae should clearly diverge from Micromonosporaceae through taxonomic classification due to inter-genera differences. However, the discovery of four novel species of Micromonosporaceae Micromonospora_E across six bacterial strains emphasizes the diversity of our marine collection, despite being so small. Currently, GTDB release 207 indicates that there are 331 Micromonosporaceae Micromonospora genomes, with two corresponding Micromonosporaceae Micromonospora_E genomes. Purely from a taxonomic perspective, we have uncovered an additional six Micromonospora_E strains that belong to a poorly represented genus in publicly available literature. Additionally, we have also discovered a total of 15 novel species of Micromonosporaceae. Even though this dataset was primarily cultivated for Micromonosporaceae strains, the taxonomic diversity acquired from collections carried out in only one marine environment hints at a dramatic and unexplored diversity in microbial NPs, clearly warranting future study.
Diversity and distribution of BGCs in marine Micromonosporaceae
The prediction, annotation and characterization of BGCs related to secondary metabolism was generated using antiSMASH v5.1.1. From the 38 Micromonosporaceae strains, 779 BGCs were annotated with a distribution of 13–34 BGCs per genome (Fig. 1). Genomes WMMA1949 (n=13), WMMA1947 (n=13) and WMMD1127 (n=13) all had the lowest number of BGCs identified, while WMMD712 (n=34) and WMMD956 (n=34) carried the highest number of BGCs identified. Manual annotation using antiSMASH-predicted BGC product types using a modified BiG-SCAPE categorization scheme to decipher non-PKS/NRPS hybrid clusters revealed a significant number of hybrid products. Across the Micromonosporaceae strains, the most heavily represented type of BGC was predicted to be involved in the production of terpenes (n=156), followed by non-PKS/NRPS hybrids (non-polyketide synthase and non-ribosomal peptide synthetase hybrids) (n=146) and RiPPs (ribosomally synthesized and post-translationally modified peptides) (n=129).
Terpenes, as the most heavily represented BGC type in our marine Micromonosporaceae, represent a structurally and functionally diverse family of natural products. Generally speaking, terpenes share a backbone chain assembly pathway that employs C5 units such as isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) as critical building blocks [7]. Terpenes displaying moderate antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) and bacteriostatic properties are well established [7]. Terpenoids also have shown antimicrobial, antifungal and anticancer cytotoxic activity [59–61]. Uncharacterized terpenes identified in our BiG-SCAPE analysis represent a largely underexploited NP grouping for possible applications against antimicrobial resistance mechanisms.
Non-PKS/NRPS hybrids were found to comprise the second largest group of BGCs in our Micromonosporaceae collection. We defined non-PKS/NRPS hybrids as antiSMASH annotated regions obeying two specific rules: (a) the region contains a hybrid of two or more products; and (b) if the hybrid contains only two products, those products must not only be a Type 1 PKS but also an NRPS. As an example, a non-PKS/NRPS hybrid could have annotations for a Type 2 PKS and RiPP or be composed of a Type 1 PKS, NRPS and terpene. This super-category was constructed to identify potential chemical hybrids that are traditionally underexplored for interesting chemistry. For instance, in WMMC264, an interleaving candidate cluster containing an NRPS, oligosaccharide and terpene overlapping was observed. NPs generally display much greater structural diversity than strictly synthetic agents and accordingly allow for much more extensive probing and utilization of chemical space; this logic is especially spurred on by the abundance of stereochemical features found in vast numbers of NPs [6]. These realizations make clear that further characterization of novel chemical spaces, as illuminated by our NP discovery efforts here, might well broaden synthetic drug approaches for target diversity [6].
RiPPs make up the third largest proportion of BGCs in our collection of Micromonosporaceae strains. RiPPs are primarily notable for being structurally diverse and displaying a wide array of biological activities [62], including potent antibiotic [63]/antifungal [64, 65]/anticancer [66, 67] activities, and expressing unique mechanisms of action compared to clinically used drugs. RiPPs are also amenable to biosynthetic engineering to mitigate systemic issues such as poor solubility and limited bioavailability [63]. Of our 779 Micromonosporaceae BGCs analysed with antiSMASH v5.1.1, 170 were found to encode a RiPP of some kind. Notably, bacteriocin (n=67), lanthipeptide (n=58), linear azol(in)econtaining peptides (n=37), thiopeptide (n=36) and Tfua-related (n=20) species comprised over 97 % of the RiPPs identified through antiSMASH (Data S13). Further characterization of these BGCs may reveal particularly malleable RiPPs with good antibiotic activities for engineering against resistant pathogens [63].
Analysis of the predicted BGCs using the BiG-SCAPE workflow revealed 328 GCFs and singletons (Fig. 2). Our analysis identified the 51 MIBiG reference BGCs as similar to our Micromonosporaceae BGCs. Specifically, BGCs known to encode the biosynthesis of gentamicin, sisomicin, rosamicin and more were linked to BGCs in our collection (Data S13). Of our 779 Micromonosporaceae BGCs, 196 BGCs (~25.1 %) clustered separately into singletons, representing a degree of uniqueness worth studying given their likelihood as beacons of unique chemical chemistry/NPs. The remaining 583 Micromonosporaceae BGCs clustered into 132 GCFs seen in Fig. 2, with 83 of the Micromonosporaceae BGCs clustering into 16 GCFs containing BGCs from MIBiG, indicating potentially known chemical compound space. The remaining 500 Micromonosporaceae BGCs clustered into 116 unique GCFs, traditionally representing unique chemical classes and indicating additional novel chemical entities potentially able to occupy unique chemical spaces. Overall, if all BGCs were sufficiently expressed in lab conditions, we would expect to have observed 312 unique chemical classes from our marine-derived Micromonosporaceae collection.
BiG-SLiCE queries reveal significant likelihood for previously unknown biosynthetic potential
The previous analysis only explored the potential chemical diversity within our Micromonosporaceae BGCs. To investigate our marine Micromonosporaceae strains’ diversity relative to published terrestrial bacteria sources, BiG-SLiCE was used as a comparative tool. BiG-SLiCE contains over ~1.2 million BGCs from publicly available literature sources, including MIBiG, which have historically been of terrestrial origin due to exhaustive mining of local ecological niches [1]. Specifically, we utilized the BiG-SLiCE model constructed using a clustering threshold value of 900, resulting in 29 955 GCFs against ~1.2 million BGCs, and queried our 779 marine Micromonosporaceae BGCs to determine individual BGC similarity to BiG-SLiCE’s GCFs (Fig. 3).
Our analysis revealed 413 BGCs that failed to cluster into the 29 955 GCF model in BiG-SLiCE (clustering threshold T=900); these BGCs thus represent distinctly novel BGCs relative to published literature, an important finding that is perhaps not that surprising given the historical bias towards terrestrial microbes reported in the literature. The remaining 366 marine-derived BGCs that successfully clustered were further categorized, informed by whether or not the closest GCF reported contains a BGC from MIBiG. From this analysis, we uncovered 94 MIBiG-associated Micromonosporaceae BGCs. Relatedly, 272 BGCs were identified as being distinct from MIBiG-associated GCFs within BiG-SLiCE. The 94 Micromonosporaceae BGCs (MIBiG-associated) probably encode known end products based on their similarity to community annotated BGCs. The remaining 272 BGCs that were not identified as MIBiG-associated may produce known end products but lack meaningful community information that might validate the exact products. In sum, the 413 BGCs that failed to cluster into the BiG-SLiCE model (T=900) represent completely unknown chemical classes, with little to no information regarding potential products beyond antiSMASH annotations. Accordingly, these BGCs showcase the probably vast biosynthetic potential of the marine environment and the microbial diversity it houses.
To contextualize our findings, we combined our BiG-SCAPE analysis and our BiG-SLiCE results to identify novel Micromonosporaceae strains that contained novel BGCs in relation to both our own dataset and to published literature. Notably, investigation of WMMD406 (Micromonosporaceae Micromonospora_E unknown sp.) in BiG-SCAPE revealed 17 BGC singletons out of the 25 total BGCs identified through antiSMASH. Comparison with BiG-SLiCE queries revealed 21 BGCs that failed to cluster (T=900) into the 29 955 GCFs that represent primarily terrestrial public literature. Of the 17 BGC singletons identified in BiG-SCAPE, 16 were shared across the 21 novel BGCs determined through BiG-SLiCE, indicating potentially unique chemical classes across our dataset and against published literature. Systematic incorporation of BiG-SCAPE analysis and BiG-SLiCE results revealed that the 413 novel BGCs identified through BiG-SLiCE made up 148 of the 196 singletons and 108 of the 132 GCFs visualized in BiG-SCAPE, representing prospective chemical classes (Data S13). Further exploration of marine Micromonosporaceae strains could continue to reveal unexplored chemical diversities and their corresponding antimicrobial activities and mechanisms.
Truly novel BGCs are identifiable using MDS and pairwise distance modelling
Our previous analyses highlighted the merits of combining BiG-SCAPE and BiG-SLiCE to identify strains and BGCs that are distinct within our dataset and also relative to published literature sources. To do this in a systematic fashion, we constructed a pairwise distance matrix (Chebyshev) of our 779 marine Micromonosporaceae BGCs using relational information regarding individual BGC distances to all 29 955 GCFs in BiG-SLiCE (T=900). Using metric MDS, we visualized similarity of our marine Micromonosporaceae BGCs against each other, split into subplots to show BGC similarity across individual BGC product types (Fig. 4). From these subplots, we observed siderophores, terpenes and RiPPs as primarily co-localizing to similar spaces based on similar GCF distance values in BiG-SLiCE. Notably, the BiG-SLiCE authors have highlighted that BGCs primarily coding for terpenes and RiPPs tend to disproportionately over-cluster due to the presence of fewer extracted features; our observations here are consistent with these earlier postulates [35]. It also is worth noting that, in the context of Fig. 4, the Hybrid (Not PKS/NRPS) category served as the positive control since it is not limited to a singular product class. Variation in this specific case was defined as the largest spread in absolute distance between BGCs defined as belonging to the same category. Manual investigation of Hybrid BGCs revealed that, in specific overlapping cases, an individual component of the Hybrid BGC was shared with the overlapping BGCs in other categories; the instances of overlap were confirmed via blast alignments. Specifically, WMMA1947 region 12, encoding putative siderophore and lanthipeptide units, showed 80.74 % identity in a blast nucleotide comparison with the siderophore subunit’s core biosynthetic gene relative to a siderophore moiety predicted to be produced by WMMD1120 region 14, also a siderophore core biosynthetic gene, seen in Data S12.
To determine how BGCs were localized following metric MDS, we scaled the marker size and hue based on the average distance of the BGC to the top three GCFs in BiG-SLiCE (Fig. 5). Further investigation into specific BGCs that were close in distance revealed that our metric MDS methodology primarily captured variation across BGC product types, but was insufficient to resolve similarity across BGCs belonging to the same predicted BGC product type. Although there is utility in comparing across BGC product types, especially when comparing BGCs that fall within the Hybrid (Not PKS/NRPS) category to other BGC predicted product categories that may share singular components to the Hybrid section, further refinements were clearly warranted.
As such, we separated our BGCxGCF dataset into sub-datasets based on the 10 predicted product types used within this paper, constructed pairwise distance matrices using the distance metrics described earlier, and performed metric MDS to enhance the power of our visualizations to resolve similar BGCs within our Micromonosporaceae (Fig. 6).
Utilizing this methodology, manual comparison with BiG-SCAPE’s SSNs and examination of antiSMASH outputs revealed that the distance between BGCs in the metric MDS visualizations corresponded strongly to the presence of shared BGC elements. For instance, WMMD975 region 20, reported in BiG-SCAPE as part of a triplet with WMMD1128 region 24 and D998 region 3, all shared a PKS/NRPS unit. Investigation of the PKS/NRPS metric MDS revealed that WMMD975 region 20 and WMMD1128 overlapped significantly, with WMMD998 region 3 distanced further away. AntiSMASH analysis (v5.1.1) revealed that WMMD975 region 20 and WMMD1128 shared the same core biosynthetic genes for the PKS and NRPS subunits, while WMMD998 contained an additional NRPS core biosynthetic gene, potentially resulting in further modifications to the prospective compound. This methodology allowed us to visually ascertain similarities across BGCs of the same predicted product type, while retaining antiSMASH-level information for the differentiation of BGC pockets.
In particular, scaling the marker size and hue based on the average distance of individual BGCs to their top three GCFs in BiG-SLiCE provided a more visually informative way to identify BGCs deviating from terrestrially derived bacteria (Fig. 7). Analysis of the PKS/NRPS category following Chebyshev-based MDS analysis revealed pockets of BGCs able to serve as representative markers of unique groups. For instance, the information regarding WMMD975’s, WMMD1128’s and WMMD998’s PKS/NRPS subunits were consistent; the average distance to the closest three GCFs in BIG-SLiCE for all BGCs exceeded 2400, indicating the likelihood of novel chemistry. Incorporation of the average distance in our methodology allowed for a visually informative way to further prioritize BGCs that are probably uncharacterized and novel compared to publicly available terrestrial data from BIG-SLiCE’s ~1.2 million BGCs (29 955 GCFs, T=900).
BGC distribution of Micromonospora E is statistically different from Micromonospora
Micromonospora_E was found to represent an underexploited region for BGCs with potentially novel chemistry. In fact, analysis of the distribution of BGC distances to the closest GCF in BiG-SLiCE uncovered a statistically significant difference between the Micromonospora and Micromonospora_E BGCs with respect to our data collection. Specifically, the mean BGC distance to the closest GCF in BiG-SLiCE for Micromonospora_E was ~1174, compared to Micromonospora’s ~1006. Similarly, the median BGC distance to the closest GCF in BiG-SLiCE showed the same trend, with Micromonospora_E reporting a median of ~1004, and Micromonospora ~908. The Mann–Whitney U-test with a two-sided hypothesis reported a P-value of 6.84e-04, less than the significance value of 0.05, indicating that the BGC distance distributions for Micromonospora and Micromonospora_E are different. In addition, analysis of the number of BGCs reporting a distance to the closest GCF in BiG-SLiCE above the threshold value (T=900), averaged over the number of strains per genera, revealed for Micromonospora a total of ~10.03 BGCs er per strain, compared to Micromonospora_E with ~14.83 BGCs per strain. Although the BiG-SLiCE pre-processed dataset was developed in 2021, access to future datasets incorporating more publicly available genomes and metagenomes will decrease the overall distances reported. However, this metric makes clear that, if we were to query a marine-derived Micromonospora strain’s BGCs against BiG-SLiCE’s currently available pre-processed dataset, we would expect to see approximately 10 BGCs that would report a distance value of ≥900, representing dissimilarity to BiG-SLiCE’s primarily terrestrial dataset, indicating potentially novel chemistry.
Conclusions
Technological advances of the last decade have had a profound impact on our ability to glimpse molecular potentials made available through the application of microbial organisms. In particular, our ability to correlate genomics information from microbes to putative small molecule products with medicinal potentials is now staggeringly advanced relative to a decade ago. With these advances has come a more complete understanding that historically employed terrestrial environments and their associated microbes may no longer warrant consideration as the optimal source of NP diversity. We provide here a comparative genomic analysis of marine Micromonosporaceae with the intention of identifying truly novel BGCs and showcasing that marine-derived organisms now represent an optimal NP source when it comes to considerations of structural and functional NP diversity. Our analyses revealed the biosynthetic diversity of marine Micromonosporaceae, especially in comparison to publicly curated databases focused on terrestrially derived genomes. Moreover, focused dissection and comparisons of the 39 individual genomic datasets chosen to showcase our bioinformatics logic allowed us to identify several novel species belonging to Micromonosporaceae Micromonospora_E. In particular, this discovery represented a 4-fold increase in known Micromonosporaceae Micromonospora_E compared to the 311 480 bacterial strains stored in GTDB-tk2 from publicly available data. Beyond the basic nuts and bolts discoveries relating marine-derived to terrestrially derived microbes and the clearly superior diversity of marine systems relative to terrestrial ones, we also endeavoured to develop a novel methodology to visually explore BGC similarities across (and within) strain collections and against publicly available data. In sum, the computational campaign disclosed here showcases: (a) the utility of prioritizing BGCs based on novelty, relative to large existing databases; and (b) the potential of renewed investigations of marine Micromonosporaceae as an outstanding repository for NP scaffolds, based largely on BGC content.
Repositories
Please note that all genome sequence data employed in this study have been deposited with NCBI. Strains and their corresponding accession numbers are as follows:
|
Strain name |
Accession (whole genome) |
Strain name |
Accession (whole genome) |
|---|---|---|---|
|
WMMA1363 |
WMMD980 |
GCA_029626035.1 |
|
|
WMMB482 |
GCA_019038635.1 |
WMMD987 |
GCA_029581435.1 |
|
WMMC250 |
GCA_027460215.1 |
WMMD937 |
GCA_029581175.1 |
|
WMMA1976 |
GCA_027497335.1 |
WMMD964 |
GCA_029581455.1 |
|
WMMA1998 |
GCA_027497375.1 |
WMMD998 |
GCA_029581235.1 |
|
WMMB486 |
WMMD1129 |
GCA_029581255.1 |
|
|
WMMA1947 |
GCA_027497355.1 |
WMMD1155 |
GCA_029581275.1 |
|
WMMA1949 |
GCA_027460145.1 |
WMMD406 |
GCA_029626025.1 |
|
WMMA2056 |
WMMD712 |
GCA_029581295.1 |
|
|
WMMA2059 |
GCA_027497315.1 |
WMMD714 |
GCA_029581315.1 |
|
WMMC241 |
GCA_027460165.1 |
WMMD718 |
GCA_029626075.1 |
|
WMMC264 |
GCA_027497275.1 |
WMMD792 |
GCA_029626105.1 |
|
WMMC273 |
GCA_027460285.1 |
WMMD956 |
GCA_029626125.1 |
|
WMMC514 |
GCA_027497295.1 |
WMMD961 |
GCA_029626145.1 |
|
WMMC500 |
GCA_027497195.1 |
WMMD967 |
GCA_029626165.1 |
|
WMMD882 |
GCA_027497255.1 |
WMMD1047 |
GCA_029626155.1 |
|
WMMD1128 |
GCA_027497235.1 |
WMMD1076 |
GCA_029581475.1 |
|
WMMD573 |
GCA_027497175.1 |
WMMD1082 |
GCA_029626175.1 |
|
WMMD710 |
GCA_029626045.1 |
WMMD1102 |
GCA_029626265.1 |
|
WMMD791 |
GCA_029581195.1 |
WMMD1120 |
GCA_029626235.1 |
|
WMMD975 |
GCA_029581215.1 |
WMMD1127 |
GCA_029626225.1 |
Funding information
This work was funded by NIH Grants U19AI109673, U19AI142720 and R01AT009874. This research was also supported by the NIGMS of the NIH under Award Number T32GM008349.
Acknowledgements
We would like to thank Professor Anthony Gitter (Dept. of Biostatistics and Medical Informatics, UWM) for suggestions regarding computational analyses. We would also like to thank the Center for High Throughput Computing for access to computational resources [68].
Author contributions
Consistent with CRediT taxonomy, the roles of contributing authors are as follows: Conceptualization: T.S.B.; Methodology: I.A., D.R.B., S.S.E., R.S.; Software: I.A., S.S.E., T.S.B.; Validation: I.A., R.S., S.S.E., T.S.B.; Formal Analysis: I.A., D.R.B., S.S.E., R.S.; Investigation: I.A., S.S.E., R.S., L.K., T.S.B.; Resources: D.R.B., S.S.E., R.S., L.K., T.S.B. ; Data Curation: I.A., S.S.E., S.R.R., T.S.B.; Writing – Original Draft: I.A., S.S.E., R.S.; Writing – Review & Editing: I.A., T.S.B.; S.R.R., T.S.B.; Visualization: I.A., S.S.E., T.S.B.; Supervision: L.K., T.S.B.; Project Administration: T.S.B.; Funding Acquisition: L.K., T.S.B.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossiter SE, Fletcher MH, Wuest WM. Natural products as platforms to overcome antibiotic resistance. Chem Rev. 2017;117:12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segala FV, Bavaro DF, Di Gennaro F, Salvati F, Marotta C, et al. Impact of SARS-CoV-2 epidemic on antimicrobial resistance: a literature review. Viruses. 2021;13:2110. doi: 10.3390/v13112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID-19: U.S Impact on Antimicrobial Resistance, Special Report 2022. National Center for Emerging and Zoonotic Infectious Diseases. 2022. [ July 10; 2023 ]. https://stacks.cdc.gov/view/cdc/117915 accessed.
- 5.Patridge E, Gareiss P, Kinch MS, Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov Today. 2016;21:204–207. doi: 10.1016/j.drudis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Stratton CF, Newman DJ, Tan DS. Cheminformatic comparison of approved drugs from natural product versus synthetic origins. Bioorg Med Chem Lett. 2015;25:4802–4807. doi: 10.1016/j.bmcl.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi S, Gui M, Li H, Yu C, Li H, et al. Secondary metabolites from marine Micromonospora: chemistry and bioactivities. Chem Biodivers. 2020;17:e2000024. doi: 10.1002/cbdv.202000024. [DOI] [PubMed] [Google Scholar]
- 8.Manivasagan P, Venkatesan J, Sivakumar K, Kim S-K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol Res. 2014;169:262–278. doi: 10.1016/j.micres.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Ellis GA, Thomas CS, Chanana S, Adnani N, Szachowicz E, et al. Brackish habitat dictates cultivable Actinobacterial diversity from marine sponges. PLoS One. 2017;12:e0176968. doi: 10.1371/journal.pone.0176968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 11.Selim MSM, Abdelhamid SA, Mohamed SS. Secondary metabolites and biodiversity of actinomycetes. J Genet Eng Biotechnol. 2021;19:72. doi: 10.1186/s43141-021-00156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramani R, Sipkema D. Marine rare actinomycetes: a promising source of structurally diverse and unique novel natural products. Mar Drugs. 2019;17:249. doi: 10.3390/md17050249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen PR, Moore BS, Fenical W. The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep. 2015;32:738–751. doi: 10.1039/c4np00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Kim S, Kim M, Lee C, Yang I, et al. Bioactive natural products from the genus Salinospora: a review. Arch Pharm Res. 2020;43:1230–1258. doi: 10.1007/s12272-020-01288-1. [DOI] [PubMed] [Google Scholar]
- 15.Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora . Proc Natl Acad Sci U S A. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letzel A-C, Li J, Amos GCA, Millán-Aguiñaga N, Ginigini J, et al. Genomic insights into specialized metabolism in the marine actinomycete Salinispora . Environ Microbiol. 2017;19:3660–3673. doi: 10.1111/1462-2920.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S, Zeng M, Wang H, Zhang H. Micromonospora: a prolific source of bioactive secondary metabolites with therapeutic potential. J Med Chem. 2022;65:8735–8771. doi: 10.1021/acs.jmedchem.2c00626. [DOI] [PubMed] [Google Scholar]
- 19.Weinstein MJ, Luedemann GM, Oden EM, Wagman GH, Rosselet JP, et al. Gentamicin,1 a new antibiotic complex from Micromonospora . J Med Chem. 1963;6:463–464. doi: 10.1021/jm00340a034. [DOI] [PubMed] [Google Scholar]
- 20.Organization WH. World Health Organization model list of essential medicines: 21st list 2019. 2019. [ October 24; 2023 ]. https://iris.who.int/handle/10665/325771 accessed.
- 21.Waksman SA, Geiger WB, Bugie E. Micromonosporin, an antibiotic substance from a little-known group of microorganisms. J Bacteriol. 1947;53:355–357. doi: 10.1128/jb.53.3.355-357.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hifnawy MS, Fouda MM, Sayed AM, Mohammed R, Hassan HM, et al. The genus Micromonospora as a model microorganism for bioactive natural product discovery. RSC Adv. 2020;10:20939–20959. doi: 10.1039/d0ra04025h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dulin CC, Sharma P, Frigo L, Voehler MW, Iverson TM, et al. EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster. J Biol Chem. 2023;299:104893. doi: 10.1016/j.jbc.2023.104893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, Zhao M, Braun DR, Ericksen SS, Piotrowski JS, et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science. 2020;370:974–978. doi: 10.1126/science.abd6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao M, Zhang F, Zarnowski R, Barns K, Jones R, et al. Turbinmicin inhibits Candida biofilm growth by disrupting fungal vesicle-mediated trafficking. J Clin Invest. 2021;131:e145123. doi: 10.1172/JCI145123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Wen Z, Liu L, Zhu X, Shen B, et al. Yangpumicins F and G, enediyne congeners from Micromonospora yangpuensis DSM 45577. J Nat Prod. 2019;82:2483–2488. doi: 10.1021/acs.jnatprod.9b00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan X, Chen J-J, Adhikari A, Yang D, Crnovcic I, et al. Genome mining of Micromonospora yangpuensis DSM 45577 as a producer of an anthraquinone-fused enediyne. Org Lett. 2017;19:6192–6195. doi: 10.1021/acs.orglett.7b03120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braesel J, Crnkovic CM, Kunstman KJ, Green SJ, Maienschein-Cline M, et al. Complete genome of Micromonospora sp. strain B006 reveals biosynthetic potential of a lake michigan actinomycete. J Nat Prod. 2018;81:2057–2068. doi: 10.1021/acs.jnatprod.8b00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics. 2022;38:5315–5316. doi: 10.1093/bioinformatics/btac672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoskisson PA, Seipke RF. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio. 2020;11:e02642-20. doi: 10.1128/mBio.02642-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terlouw BR, Blin K, Navarro-Muñoz JC, Avalon NE, Chevrette MG, et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023;51:D603–D610. doi: 10.1093/nar/gkac1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023;51:W46–W50. doi: 10.1093/nar/gkad344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kautsar SA, van der Hooft JJJ, de Ridder D, Medema MH. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience. 2021;10:giaa154. doi: 10.1093/gigascience/giaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saati-Santamaría Z, Selem-Mojica N, Peral-Aranega E, Rivas R, García-Fraile P. Unveiling the genomic potential of Pseudomonas type strains for discovering new natural products. Microb Genom. 2022;8:000758. doi: 10.1099/mgen.0.000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyche TP, Hou Y, Braun D, Cohen HC, Xiong MP, et al. First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J Org Chem. 2011;76:6542–6547. doi: 10.1021/jo200661n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, et al. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, Keyicin. ACS Chem Biol. 2017;12:3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molecular Biology and Evolution, Oxford Academic; [ October 24; 2022 ]. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes.https://academic.oup.com/mbe/article/38/10/4647/6329644 n.d. accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alanjary M, Kronmiller B, Adamek M, Blin K, Weber T, et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017;45:W42–W48. doi: 10.1093/nar/gkx360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mungan MD, Alanjary M, Blin K, Weber T, Medema MH, et al. ARTS 2.0: feature updates and expansion of the antibiotic resistant target seeker for comparative genome mining. Nucleic Acids Res. 2020;48:W546–W552. doi: 10.1093/nar/gkaa374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waskom ML. seaborn: statistical data visualization. JOSS. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 47.Van Rossum G, Drake FL. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- 48.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE V5 enables improved estimates of phylogenetic tree confidence by ensemble bootstrapping. bioRxiv. 2021:2021. [Google Scholar]
- 50.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 55.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cantrell CD. Modern Mathematical Methods for Physicists and Engineers. Cambridge University Press; 2000. [DOI] [Google Scholar]
- 59.Guimarães AC, Meireles LM, Lemos MF, Guimarães MCC, Endringer DC, et al. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules. 2019;24:2471. doi: 10.3390/molecules24132471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomko AM, Whynot EG, Ellis LD, Dupré DJ. Anti-cancer potential of Cannabinoids, Terpenes, and Flavonoids present in Cannabis. Cancers. 2020;12:1985. doi: 10.3390/cancers12071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao A, Zhang Y, Muend S, Rao R. Mechanism of antifungal activity of terpenoid phenols resembles calcium stress and inhibition of the TOR pathway. Antimicrob Agents Chemother. 2010;54:5062–5069. doi: 10.1128/AAC.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scherlach K, Hertweck C. Mining and unearthing hidden biosynthetic potential. Nat Commun. 2021;12:3864. doi: 10.1038/s41467-021-24133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudson GA, Mitchell DA. RiPP antibiotics: biosynthesis and engineering potential. Curr Opin Microbiol. 2018;45:61–69. doi: 10.1016/j.mib.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hetrick KJ, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr Opin Chem Biol. 2017;38:36–44. doi: 10.1016/j.cbpa.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohr KI, Volz C, Jansen R, Wray V, Hoffmann J, et al. Pinensins: the first antifungal lantibiotics. Angew Chem Int Ed Engl. 2015;54:11254–11258. doi: 10.1002/anie.201500927. [DOI] [PubMed] [Google Scholar]
- 66.Russell AH, Truman AW. Genome mining strategies for ribosomally synthesised and post-translationally modified peptides. Comput Struct Biotechnol J. 2020;18:1838–1851. doi: 10.1016/j.csbj.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frattaruolo L, Lacret R, Cappello AR, Truman AW. A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem Biol. 2017;12:2815–2822. doi: 10.1021/acschembio.7b00677. [DOI] [PubMed] [Google Scholar]
- 68.Center for High Throughput Computing. 2006. https://chtc.cs.wisc.edu/