Fig. 2 |. oRPs interact with Sis1/DnaJB6 at the nucleolar periphery.
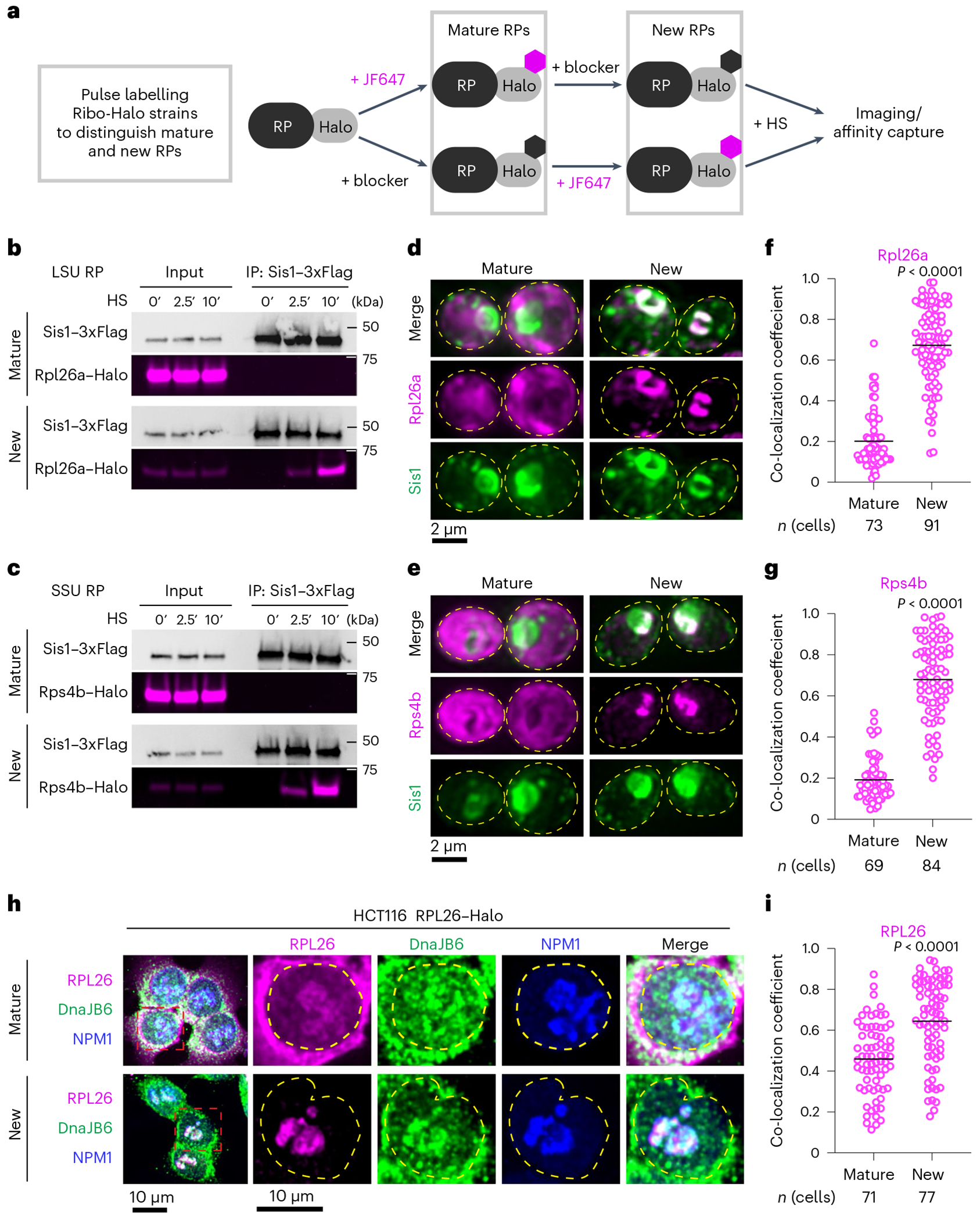
a, Workflow for in vivo pulse-labelling of mature and new RPs. b, IP of Sis1–3xFlag and either mature or new Rpl26a–Halo from cells left unstressed or heat shocked at 39 °C for the indicated times. n = 2 biologically independent experiment. LSU, 60S large subunit. c, As in b, but for Rps4b–Halo. n = 2 biologically independent experiment. SSU, 40S small subunit. d, Lattice light sheet live imaging of yeast under heat shock (39 °C, 10 min) expressing Sis1–mVenus and labelled for either new or mature Rpl26a–Halo. The dashed line indicates the cellular boundary. e, As in c, but for Rps4b–Halo. f, Co-localization (Mander’s overlap coefficient) of Sis1–mVenus with either mature or new Rpl26a–Halo in heat-shocked cells (39 °C, 10 min). P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from four independent biological replicates. g, As in f, but for Rps4b–Halo. P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from four independent biological replicates. h, Human HCT116 cells stably expressing RPL26–Halo labelled for mature or new RPL26 and heat shocked (43 °C for 30 min). Cells were fixed and immunostained for DnaJB6 and NPM1. Dashed line indicates the nuclear boundary. i, Co-localization (Mander’s overlap coefficient) of DnaJB6 with either mature or new RPL26–Halo in heat-shocked cells (43 °C, 30 min). P values were calculated with unpaired two-tailed Welch’s t-test. n indicates number of cells, pooled from three independent biological replicates.
