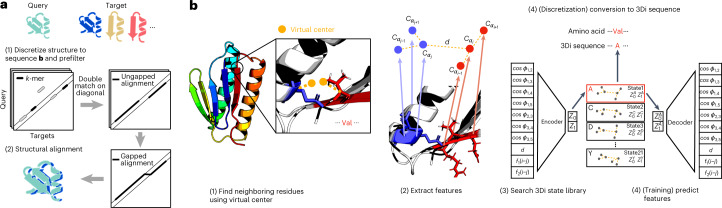
Fig. 1. Foldseek workflow.
a, Foldseek searches a set of query structures through a set of target structures. (1) Query and target structures are discretized into 3Di sequences (see b). To detect candidate structures, we apply the fast and sensitive k-mer and ungapped alignment prefilter of MMseqs2 to the 3Di sequences, (2) followed by vectorized Smith–Waterman local alignment combining 3Di and amino acid substitution scores. Alternatively, a global alignment is computed with a 1.7-times accelerated TM-align version (Supplementary Fig. 12). b, Learning the 3Di alphabet. (1) 3Di states describe tertiary interaction between a residue i and its nearest neighbor j. Nearest neighbors have the closest virtual center distance (yellow). Virtual center positions (Supplementary Fig. 1) were optimized for maximum search sensitivity. (2) To describe the interaction geometry of residues i and j, we extract seven angles, the Euclidean Cα distance and two sequence distance features from the six Cα coordinates of the two backbone fragments (blue and red). (3) These 10 features are used to define 20 3Di states by training a VQ-VAE28 modified to learn states that are maximally evolutionary conserved. For structure searches, the encoder predicts the best-matching 3Di state for each residue.