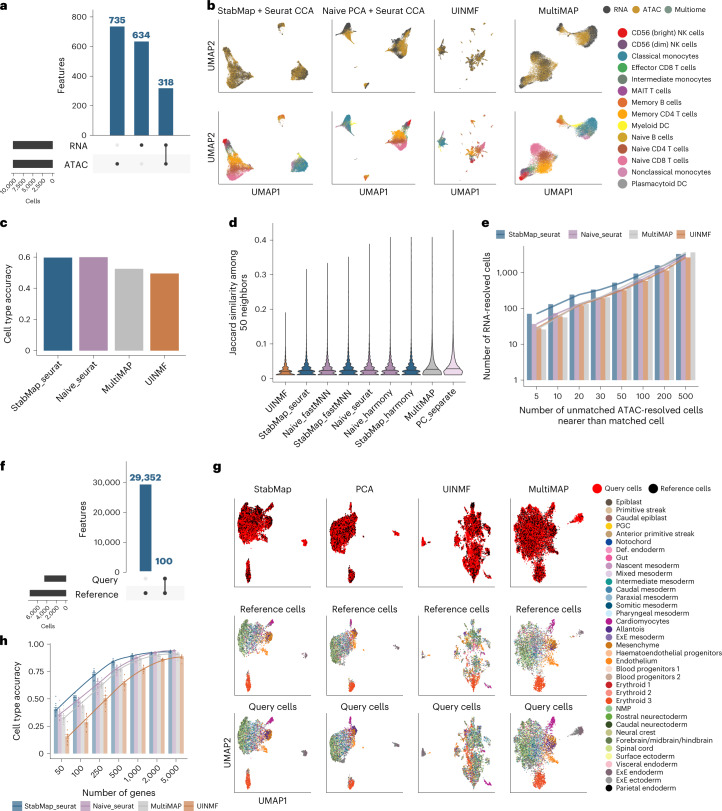
Fig. 2. Mosaic data integration simulations using PBMC Multiome and Mouse Gastrulation Atlas data.
a, UpSet plot of features shared between simulated RNA and ATAC modalities. ATAC peaks in promoter regions of genes are aligned with the genes in the RNA modality, resulting in 318 common features, 735 and 634 features distinct to the ATAC and RNA platforms, respectively. b, UMAP representations of RNA and ATAC modality cells for StabMap (first column), PCA, UINMF and MultiMAP (last column), colored by simulated modality (top row) and by cell type (bottom row). c, Bar plot of cell type classification accuracy predicting ATAC-resolved cell types using RNA-resolved cells as training data. d, Violin plots displaying Jaccard similarity among 50 neighbors for cells in each modality, where a higher value indicates a better preservation of neighborhood structure. e, Bar plot displaying the cumulative number of RNA-resolved cells, grouped by the number of unmatched ATAC-resolved cells found to be nearer than the matched ATAC-resolved cell. Ideally all RNA-resolved cells would be placed near their matching ATAC-resolved cells; therefore, more positive values indicate more cells nearer to their true matching cell and better quality of recapturing cell relationships. f, UpSet plot of features between simulated query and reference datasets for Mouse Gastrulation Atlas data. In this example the query dataset contains only 200 features, whereas the reference dataset contains those features along with 9,372 additional features. g, UMAP representations of Mouse Gastrulation Atlas data simulation scenario described in f using StabMap, PCA, MultiMAP and UINMF. The first row shows the query cells colored by cell type, the second row shows reference cells colored by cell type, and the third row shows query cells colored by cell type. h, Bar plot displaying the cell type classification accuracy of query cells for various methods, when the query set is restricted to different numbers of genes. Error bars represent mean ± standard error of the mean. Cell type classification is performed for all combinations of query and reference sample sets, totaling 12 repetitions. Def. endoderm = definitive endoderm. ExE mesoderm = extraembryonic mesoderm.