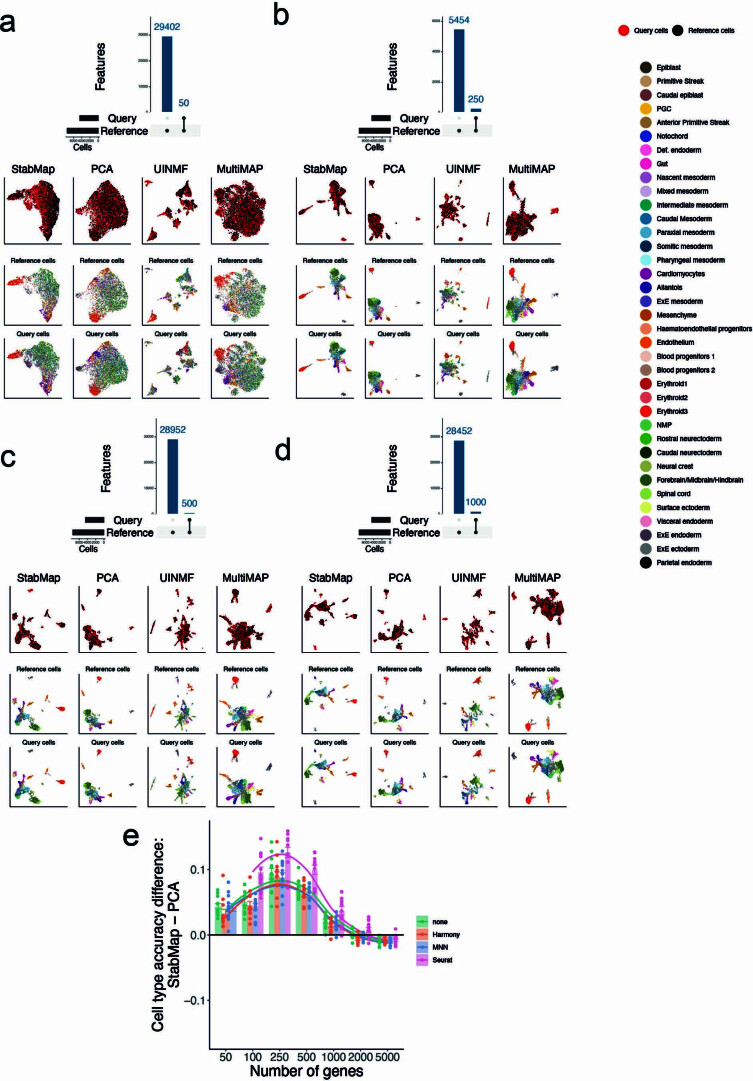
Extended Data Fig. 1. Comparison of StabMap using Mouse Gastrulation Atlas data.
a. UpSet plot and UMAP representations of Mouse Gastrulation Atlas data simulation with 100 randomly selected features using StabMap, PCA, MultiMAP, and UINMF. First row shows the query cells coloured by simulated dataset, the second row shows reference cells coloured by cell type, and the third row shows query cells coloured by cell type. b-d. As in panel (a.) for 500, 1,000, randomly selected and all features respectively. e. Barplot displaying the difference in cell type prediction accuracy (y-axis) in the Mouse Gastrulation Data simulation, where data is integrated using StabMap or the naive PCA approach. StabMap displays a higher cell type accuracy for many choices of the number of genes (x-axis) for all choices of downstream horizontal integration (none, Harmony, Mutual Nearest Neighbours (MNN) and Seurat), and as the number of genes increases, this difference reduces closer to zero, indicating that the gain in accuracy is much more pronounced for smaller numbers of genes. Cell type classification is performed for all combinations of query and reference sample sets totalling 12 repetitions. Data are presented as mean values + /- SEM.