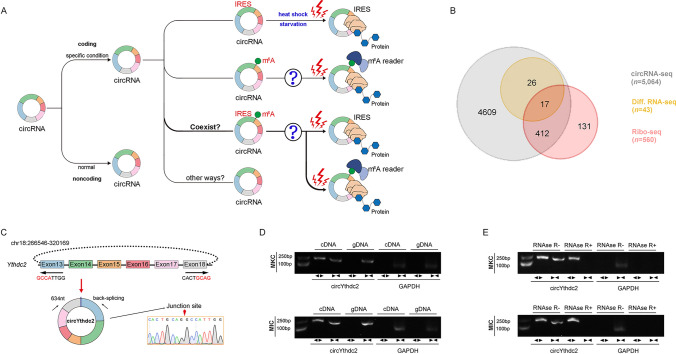
Fig. 1.
Expression profiles and characterization of circYthdc2. A Schematic diagram of circRNA translation ways and its selection conditions. B Strategies used for circRNA-seq and ribosome profiling (Ribo-seq). The gray strips represented the total circRNAs by circRNA-Seq. The orange strips represented the differential circRNAs upon SCRV treatment. The red strips represented the circRNAs with potential translation ability by Ribo-seq. C We confirmed the head-to-tail splicing of circYthdc2 in the circYthdc2 RT-PCR product by Sanger sequencing. D RT-PCR validated the existence of circYthdc2 in MKC and MIC cell lines. CircYthdc2 was amplified by divergent primers in cDNA but not gDNA. GAPDH was used as a negative control. E The expression of circYthdc2 and linear Ythdc2 mRNA in both MKC and MIC cell lines was detected by RT-PCR assay followed by nucleic acid electrophoresis or qPCR assay in the presence or absence of RNase R. All data represented the three independent triplicated experiments