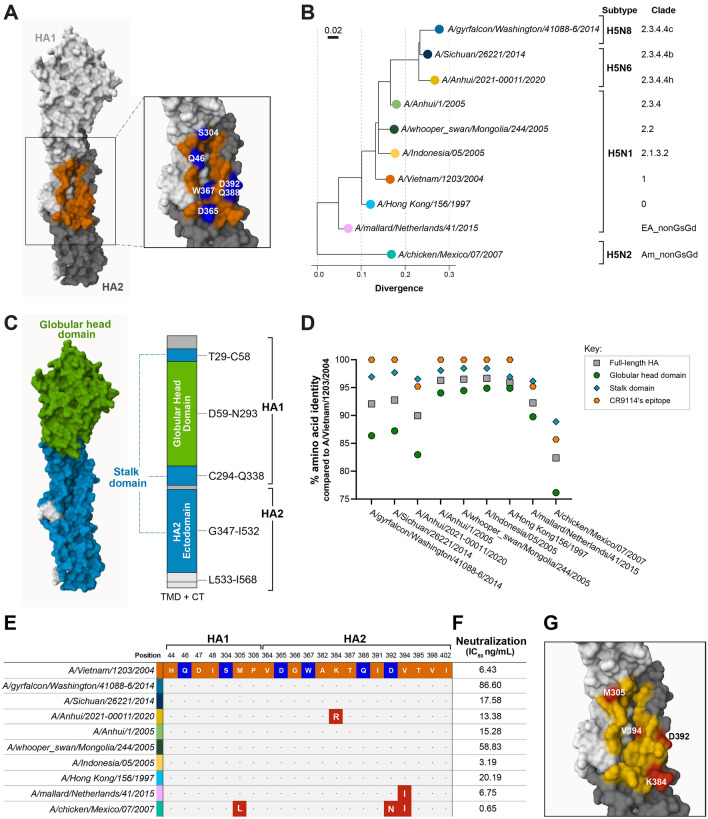
Figure 1.
CR9114 neutralizes H5 strains belonging to different clades in vitro due to the high level of conservation of its epitope, and even in case of epitope substitutions. (A) CR9114’s epitope on A/Vietnam/1203/2004 HA16. The surfaces of HA1 and HA2 subunits are shown in white and grey, respectively. Residues in the epitope are shown in orange, while hotspots, defined as residues forming H-bonds with CR9114, are shown in blue. The numbering of the residues follows the amino acid sequence alignment shown in Supplementary Fig. S1. (B) Phylogenetic tree based on the identity of the hemagglutinin protein sequence, showing the divergence among the strains used in the pseudotype virus neutralization assay (PNA). The selected strains belong to different clades. A/chicken/Scotland/1959 (not shown) was used as a root for building the tree. The full tree is presented in Supplementary Fig. S2. The list of details regarding the H5 strains is reported the Supplementary Table S1. (C) Schematic depiction of the full-length hemagglutinin from A/Vietnam/1203/2004 HA showing the boundaries of the globular head and of the stalk domains. The stalk domain consists of the N- and C-terminal parts of HA1 subunit, and the HA2 ectodomain; the transmembrane domain (TMD), and the cytoplasmic tail (CT) on the HA2 are not shown on the 3D structure. (D) Percent identity of amino acid sequence of the strains tested in the PNA compared to the full-length HA, globular head domain, stalk domain, and CR9114’s epitope of A/Vietnam/1203/2004. (E) Sequence alignment of CR9114’s epitope on H5 HA from A/Vietnam/1203/2004 and the other strains used in the PNA. Hot spots are highlighted in blue. (F) Neutralization values as IC50 values (ng/mL) generated by PNA of CR9114 against ten H5 strains belonging to difference clades. (G) CR9114’s epitope on the HA of the strains with amino acid substitutions compared to A/Vietnam/1203/2004 tested in the PNA. Residues in yellow are fully conserved, whereas residues in red have been substituted in at least one of the tested strains. Figure 1A, C and G have been generated in Mol* Viewer19 with PDB ID: 4FQI. Single-letter abbreviations for amino acids: A Ala, D Asp, G Gly, H His, I Ile, K Lys, L Leu, M Met, N Asn, P Pro, Q Gln, S Ser, T Thr, V Val, W Trp.