Figure 3.
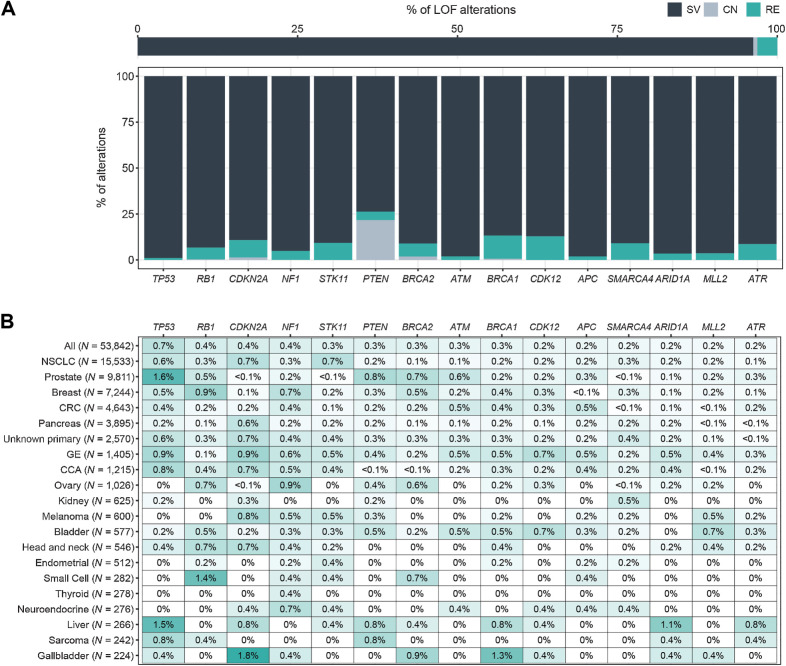
LOF rearrangements. A, Relative prevalence of short variants (SV), copy-number deletions (CN), and rearrangements (RE) predicted to disrupt tumor suppressor genes: for all tumor suppressor genes (top), and for the top 15 rearranged tumor suppressor genes (bottom). CN is only reported for PTEN and BRCA1/2. B, Heatmap of the prevalence of the most frequently disrupted tumor suppressor genes in the pan-tumor cohort. Cancer types with ≥200 LBx and genes altered in >0.5% of at least 1 cancer type are shown.