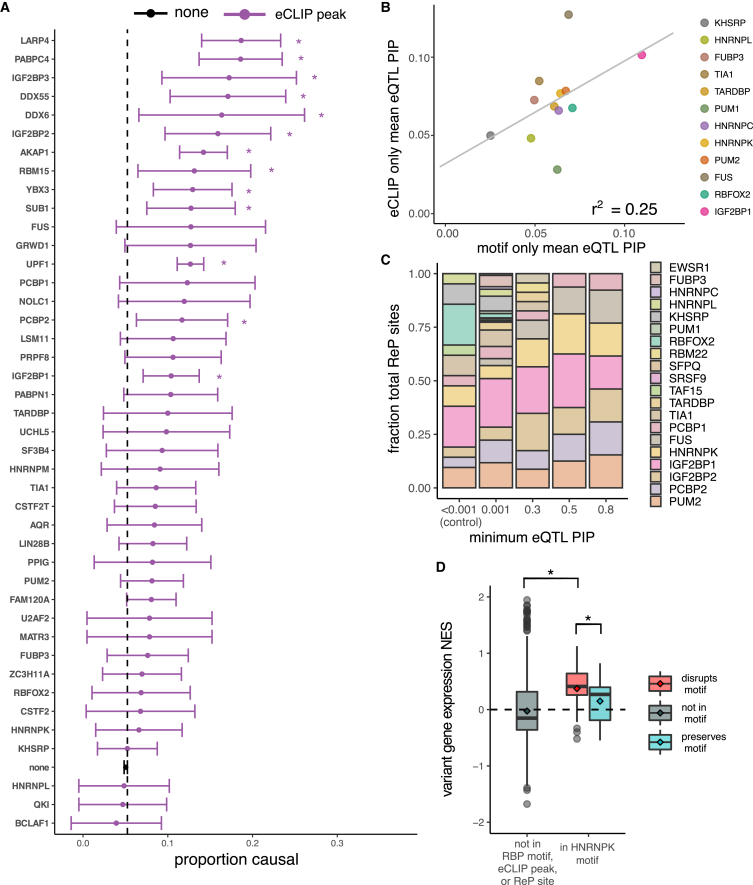
Figure 3.
Certain RBP motifs and eCLIP peaks are enriched among eQTLs and alter expression
(A) Proportion causal (with 95% confidence interval) for variants in eCLIP peaks for different RBPs, for all RBPs with ≥50 variants in eCLIP peaks. The proportion causal for variants not in any eCLIP peak was 0.05 (dashed line). ∗p < 0.01.
(B) Mean PIP for eQTLs in eCLIP peaks but not RBP motifs (y axis) versus mean PIP for eQTLs in RBP motifs but not eCLIP peaks, for all RBPs in both datasets. Shown is the regression line with Pearson correlation coefficient.
(C) Distribution of RBPs among ReP sites at different minimum eQTL PIP cutoffs. Shown are RBPs representing at least 0.1% of all ReP sites.
(D) Variant NES (from GTEx) on gene expression for high-confidence (PIP > 0.9) eQTLs not in RBP motifs or eCLIP peaks (gray), and for high-confidence eQTLs predicted to disrupt (red) or preserve (blue) HNRNPK motifs. ∗p < 0.01.