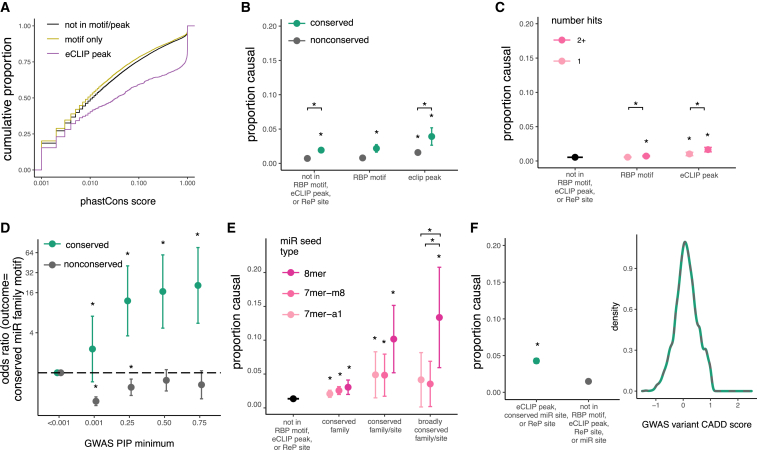
Figure 4.
3′ UTR variants in RBP motifs, eCLIP peaks, and miRNA sites are associated with phenotypes
(A) Comparison of PhastCons score distributions for GWAS variants in eCLIP peaks, RBP motifs, or no known regulatory elements.
(B) Proportion causal (proportion of GWAS hits with PIP > 0.25) with 95% confidence intervals for variants not in RBP motifs or eCLIP peaks compared to variants in RBP motifs or eCLIP peaks.
(C) As in (B), but for variants in a single motif or CLIP peak compared to variants in more than one motif or peak in genes matched by gene expression.
(D) Odds of a GWAS variant being in predicted miRNA site versus control variants (PIP < 0.001) as minimum PIP increases; shown is odds ratio with 95% confidence intervals.
(E) Proportion causal with 95% confidence intervals for variants not in miRNA sites compared to variants in miRNA sites with increasing predicted seed strength.
(F) Proportion causal with 95% confidence intervals (left) for variants not in RBP motifs or eCLIP peaks compared to variants in ReP sites or eCLIP peaks, matched for raw CADD score (right).
For all panels, ∗p < 0.01.