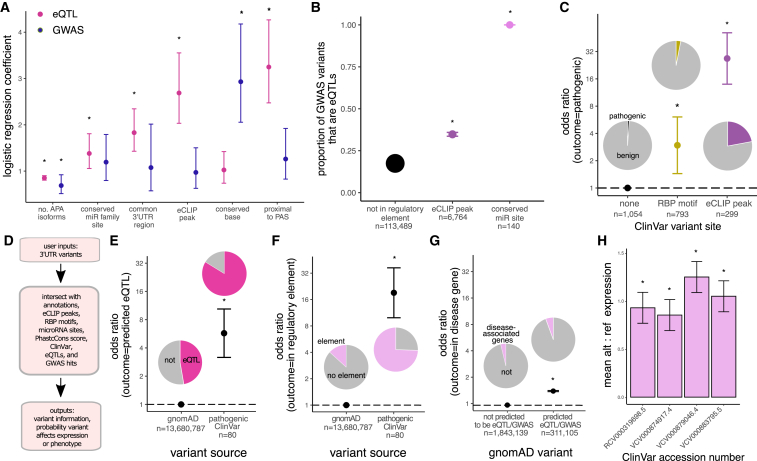
Figure 6.
Characterization of 3′ UTR variants into their annotations and regulatory elements helps prioritize variants for functional analysis and disease classification
(A) Exponential of logistic regression model coefficients with 95% confidence intervals. The model independent variable is binary (PIP greater or less than/equal to 0.5), and ∗p < 0.05.
(B) Intersection of eQTL variants in different putative regulatory elements with GWAS hits; dots are weighted by intersect size and error bars are 95% confidence intervals.
(C) Odds of a ClinVar 3′ UTR variant in RBP motifs or eCLIP peaks being pathogenic versus variants not in a predicted regulatory element; shown is odds ratio with 95% confidence intervals.
(D) RegVar workflow and example output for two ClinVar 3′ UTR variants of uncertain clinical significance (genomic coordinates are in hg38).
(E) Odds of a pathogenic ClinVar 3′ UTR variant being predicted by RegVar to be an eQTL versus all 3′ UTR gnomAD variants, with 95% confidence intervals.
(F) Odds of a pathogenic ClinVar 3′ UTR variant being in a predicted regulatory element (miRNA site, RBP motif, or eCLIP peak) versus all 3′ UTR gnomAD variants, with 95% confidence intervals.
(G) Odds of a gnomAD 3′ UTR variant predicted by RegVar to be an eQTL to be in a disease-associated gene versus 3′ UTR gnomAD variants not predicted to be eQTLs, with 95% confidence intervals.
(H) Difference in reference versus alternative allele reporter RNA expression in cell lines for select ClinVar variants. Confidence intervals are standard deviation of two technical replicates.
For all panels except (A), ∗p < 0.01.