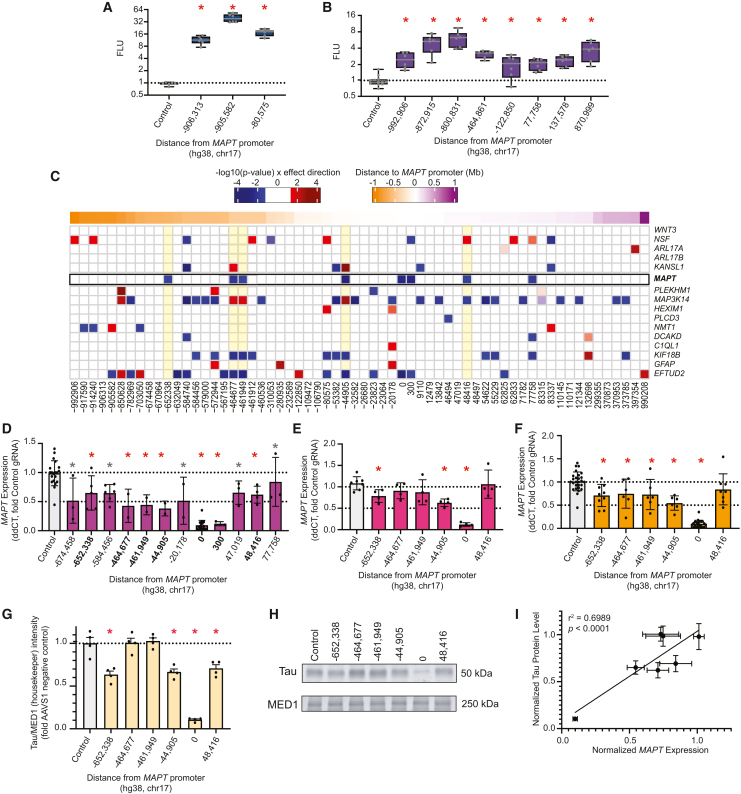
Figure 4.
Functional validation of nominated CREs
(A) Boxplots showing statistically significant (∗p < 0.05, ANOVA with Fisher’s LSD) elements tested in luciferase assays in iCell GlutaNeurons.
(B) Boxplots showing statistically significant (∗p < 0.05, ANOVA with Fisher’s LSD) elements tested in luciferase assays in KOLF2.1J-hNGN2 neurons.
(C) Heatmap showing differential expression following CRISPRi experiments in day 14 XCL4 BrainPhys differentiated neurons of MAPT and all genes expressed (cpm > 30) within 1 Mb of MAPT TSS. X axis indicates the distance of the midpoint of the target region tested in CRISPRi experiments from the MAPT promoter (chr17:45,894,000).
(D) RT-qPCR of MAPT expression. Barplots show statistically significant regions tested. Red asterisk indicates regions significant by 3′ mRNA-seq and RT-qPCR. Gray asterisk indicates regions significant in either RT-qPCR or 3′ mRNA-seq. ∗p < 0.05, ANOVA with Fisher’s LSD.
(E) Replication of five key regions identified in (C) and (D) in KOLF2.1J BrainPhys differentiated neurons.
(F) Meta-analysis combining RT-qPCR of resulting neuronal MAPT expression data from both the XCL4 and KOLF2.1J BrainPhys neurons.
(G) Quantification of western blot (∗p < 0.05 by ANOVA with a Dunnett’s multiple comparisons post-hoc vs. AAVS1 safe harbor control).
(H) Western blot of Tau and MED1 (housekeeper) protein levels following CRISPRi perturbation.
(I) Correlation plot of MAPT RNA transcript abundance with tau protein level using simple linear regression analysis.