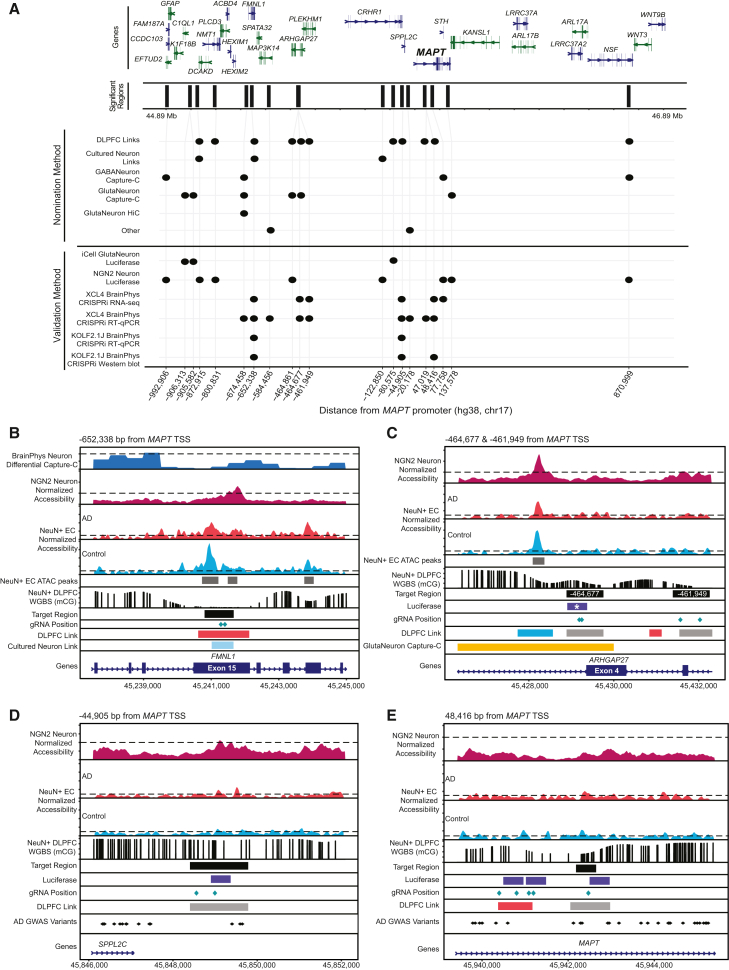
Figure 5.
Overview of regions with sufficient evidence of CRE activity
(A) Browser view of significant nominated regions by functional assay. Top: gene model of genes within the locus (hg38, chr17: 44.89–46.89 Mb). Middle: regions with significant activity in functional assays denoted by their distance from the MAPT TSS. Bottom: upset plot showing functional assays in which cCREs had significant activity.
(B–E) Key CREs regulating MAPT expression. Zoom in on regions validated by qPCR and CRISPRi in XCL4 BrainPhys neurons (B) −652,338, (C) −464,677 and −461,949, (D) −44,905, and (E) 48,416. Top panels: differential Capture-C in BrainPhys Neurons (range 0–2), normalized accessibility from KOLF2.1J-hNGN2 differentiated Neurons (range 0–6), and normalized accessibility from NeuN+ entorhinal cortex (EC) postmortem tissue from individuals with AD (red) and healthy control subjects (blue, range 0–20). Called peaks are shown as gray bars. Bottom panels: NeuN+ DLPFC methylated CpGs from whole-genome bisulfite sequencing (WGBS, range 0–1). Tracks of target region, luciferase region (∗p < 0.05, ANOVA with Fisher’s LSD), gRNA position, significant AD GWAS variants, and nominated regions (DLPFC Links: blue, control-specific link; gray, common link; red, AD-specific link).