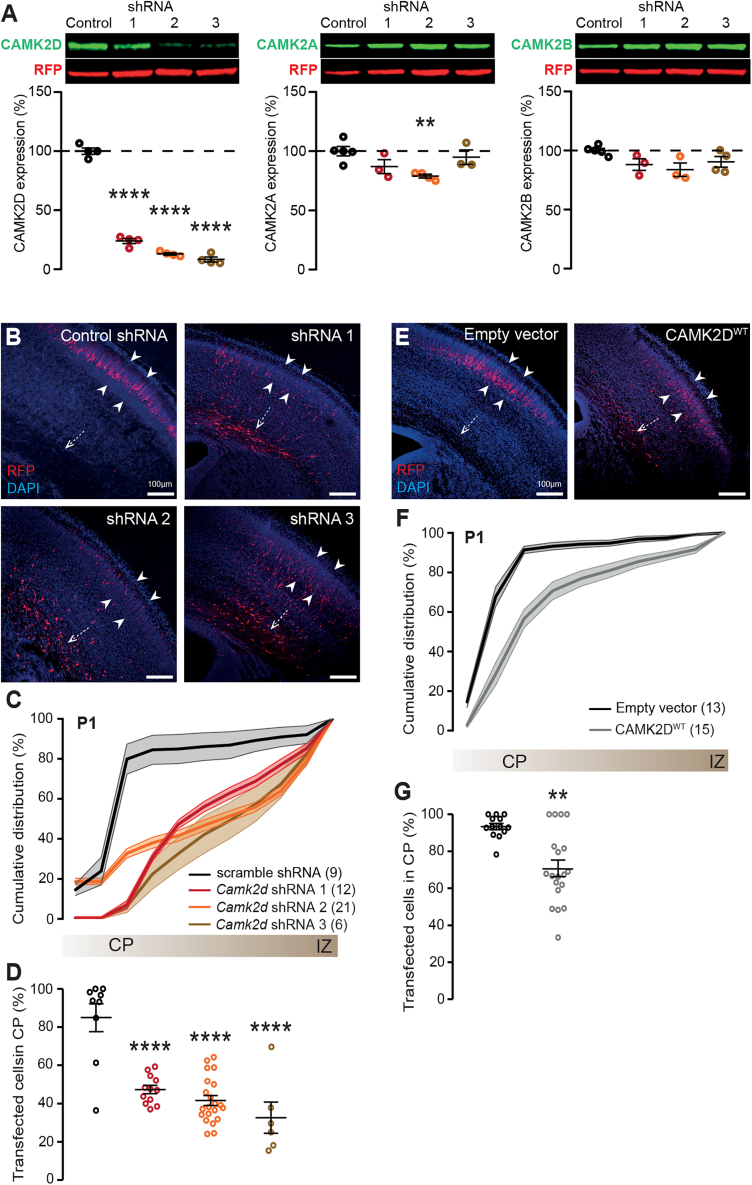
Figure 1.
Reduced as well as increased CAMK2D protein level during prenatal neurodevelopment results in a migration delay
(A) Knockdown of Camk2d with shRNAs proves to be efficient and mostly specific. Top: representative western blots of protein lysates from HEK293T cells, which were transfected with either control (scramble) or Camk2d specific shRNA’s. Blots were probed with an antibody against the specific CAMK2 (green), and RFP (red). Bottom: Quantification of CAMK2D (left), CAMK2A (middle), or CAMK2B (right) protein levels normalized against RFP.
(B) Representative images of coronal brain slices from P0/P1 pups that were transfected with scramble or Camk2d-specific shRNAs at E14.5, using in utero electroporation. White dashed arrow indicates the subventricular zone (SVZ); white arrowheads indicate the cortical plate (CP). DAPI is in blue, RFP in red.
(C) Cumulative graph indicating the migration pattern from the SVZ to the CP in presence of scramble or Camk2d-specific shRNAs.
(D) Quantification of tdTomato-positive cells that have successfully migrated to the CP, revealing that knockdown of Camk2d leads to a delay in migration.
(E) Representative images of coronal brain slices from P0/P1 pups that were transfected with a control empty vector or CAMK2DWT at E14.5, using in utero electroporation.
(F) Cumulative graph indicating the migration pattern from the SVZ to the CP upon overexpression of the empty vector or CAMK2DWT.
(G) Quantification of tdTomato-positive cells that have successfully migrated to the CP, showing that overexpression of CAMK2DWT leads to a delay in migration as well. Number in parentheses represents the number of images used for the quantification; dots represent data points and error bars indicate SEM; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.