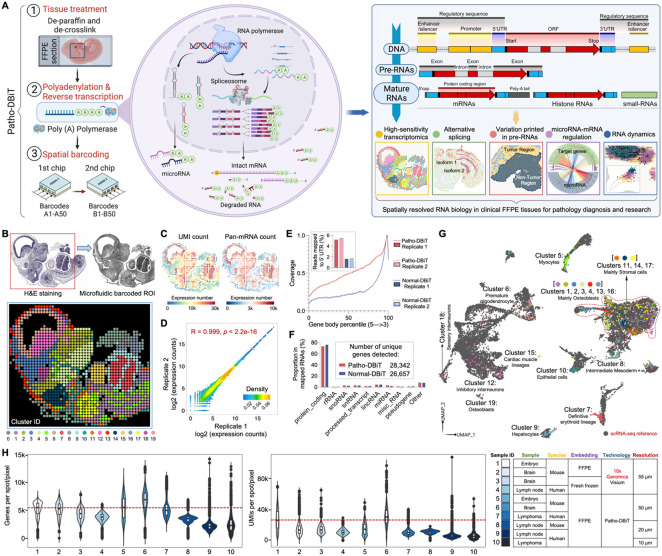
Figure 1. Patho-DBiT workflow, technical performance, and spatial mapping of mouse embryo.
(A) Schematic workflow, molecular underpinnings, and technological spectrum of Patho-DBiT. Three major steps include (1) FFPE tissue de-paraffinization and de-crosslink, (2) Enzymatic in situ polyadenylation and reverse transcription, (3) Spatial barcoding using a pair of microfluidic devices. Patho-DBiT utilizes poly(A) polymerase to add poly(A) tails to both A-tailed intact mRNA and non-A-tailed RNAs, enabling spatial characterization of molecules across the entire transcription process. Patho-DBiT demonstrates spatial profiling of high-sensitivity transcriptome, alternative splicing, variations printed in pre-RNAs, microRNAs, and RNA dynamics.
(B) Patho-DBiT's performance and versatility on an E13 mouse embryo FFPE section. Top left: H&E staining of an adjacent section. Red square indicates the region of interest (ROI). Top right: tissue scanning post 50μm-microfluidic device barcoding. Bottom: unsupervised clustering identified 20 transcriptomic clusters, closely aligning with the H&E tissue histology.
(C) Spatial pan-mRNA and UMI count maps.
(D) Correlation analysis between replicates shows the high reproducibility of Patho-DBiT. Pearson correlation coefficient is indicated.
(E) Read coverage along the gene body from 5' to 3’ and the percentage of reads mapped to the 5' UTR. Comparison involves two Patho-DBiT replicates with normal DBiT mapping without polyadenylation.
(F) Comparison of the proportion of mapped RNA categories between Patho-DBiT and normal DBiT. Patho-DBiT demonstrates a similarly low level of mapped rRNA percentage compared to normal DBiT.
(G) Integration of spatial RNA data with scRNA-seq mouse organogenesis data (Cao et al., Nature 2019).
(H) Distribution of gene and UMI counts in different tissue types at varying spatial resolutions. Patho-DBiT is benchmarked against another sequencing-based spatial technology, Visium from 10x Genomics on both FFPE and fresh frozen tissues.