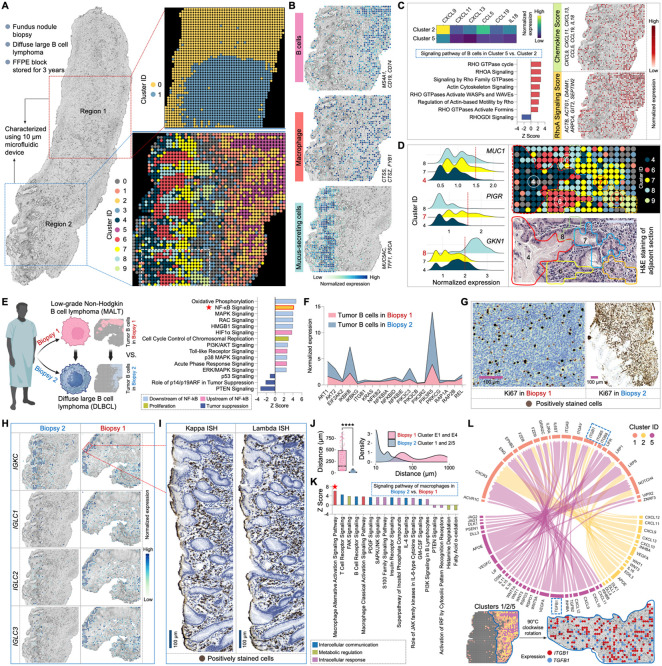
Figure 7. Cellular level spatial mapping of a DLBCL section elucidates tumor progression.
(A) Spatial transcriptome mapping of fundus nodule biopsy sections collected from the same patient depicted in Figure 4 (A) at the same time. The diagnosis progressed from low-grade MALT to DLBCL in this subsequent biopsy. Left: sections from two different regions underwent 10μm-microfluidic device spatial barcoding. Right top: unsupervised clustering of Region 1 identified two clusters. Right bottom: unsupervised clustering of Region 2 revealed 10 transcriptomically distinct subpopulations.
(B) Spatial characterization of representative cell types based on the expression of signature gene. Genes defining each module score are listed.
(C) Spatial heterogeneities and interactions among tumor B cells. Left top: comparative analysis of chemokine gene expression between clusters 2 and 5. Left bottom: signaling pathways regulated by DEGs between cluster 2 vs. cluster 5. Right: spatial distribution of the Chemokine Score and RhoA Signaling Score. Genes defining each module score are listed.
(D) Cellular-level spatial mapping unveils a distinct transcriptomic neighborhood. Left: comparative analysis of gastric mucus-secreting cell related gene expression between clusters 4, 7, and 8. Right top: enlarged transcriptomic neighborhood highlighted by white square in (A). Right bottom: tissue morphology of the corresponding area defined by H&E staining of an adjacent section.
(E) Spatial analysis elucidates the molecular dynamics driving tumor progression. Left: schematic illustration showing comparative analysis. Right: signaling pathways regulated by DEGs between tumor B cells in DLBCL vs. MALT biopsy, revealing a significant upregulation of NF-κB signaling and its associated upstream and downstream pathways.
(F) Expression comparison of key genes involved in the NF-κB signaling between DLBCL vs. MALT biopsy.
(G) IHC staining for Ki67 on adjacent sections from the two biopsies.
(H) Spatial expression mapping of genes encoding plasma cell kappa and lambda chains in the two biopsies.
(I) ISH staining for kappa and lambda chain mRNA in the designated area in (H).
(J) Distance distribution between macrophages and tumor B cells in the two biopsies. Significance level was calculated with two-tailed Mann-Whitney test, **** P < 0.0001.
(K) Signaling pathways regulated by DEGs between macrophages in DLBCL vs. MALT biopsy, revealing a significant upregulation of macrophage alternative activation signaling and its associated pathways.
(L) Ligand-receptor interactions between macrophage cluster 1 and tumor B cell clusters 2 and 5. The distinctive communication pattern of TGF-β (TGFB1) and the integrin family (ITGB1, ITGB5, and ITGB8) is indicated and spatially visualized. Edge thickness is proportional to correlation weights.
In (C), (E) and (K), z score is computed and used to reflect the predicted activation level (z>0, activated; z<0, inhibited; z≥2 or z≤−2 can be considered significant).