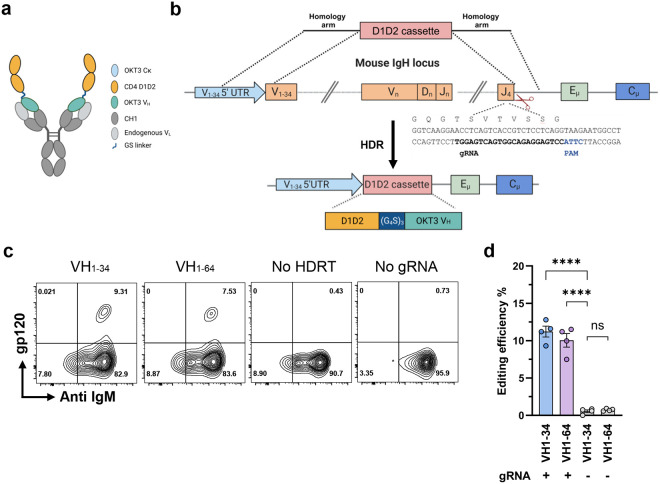
Fig. 1. Engineering primary murine B cells to express a B-cell receptor with CD4 domains 1 and 2.
a A representation of an engineered BCR with a potency and half-life enhanced form of CD4 domains 1 and 2 (D1D2) fused through a (G4S)3 linker to the amino-terminus of the heavy-chain variable region of the mouse antibody OKT3 (D1D2-OKT3-VH). The OKT3 heavy chain pairs with an endogenous mouse light chain. b Introducing D1D2-OKT3-VH at the murine heavy-chain locus. The CRISPR effector protein Mb2Cas12a targets the J4 coding region 5’ of a CTTA PAM, as represented. An rAAV-delivered homology directed repair template (HDRT) complements the 5’ UTR of a VH segment and the intron 3’ of JH4 using 576 bp and 600 bp homology arms, respectively. The edited genome replaces the VDJ-recombined heavy chain with a cassette encoding D1D2-OKT3-VH. c Expression of D1D2-OKT3-VH in primary mouse B cells. Expression of D1D2 in edited cells was measured by flow cytometry with monomeric HIV-1 gp120. Representative flow cytometry plots of B cells edited with HDRT targeting the 5’ UTR of VH1–34 or V1–64 were generated 48 h after electroporation. HDRT were delivered with rAAV transduced at 104 multiplicity of infection (MOI). Controls include cells electroporated with Mb2Cas12a ribonucleoproteins (RNP) without rAAV (No HDRT) or without gRNA but transduced with HDRT-encoding rAAV (No gRNA). Plots were gated on viable singlet B cells. d Quantitation of editing efficiency in c from independent experiments. Each dot represents an average from two biologically independent replicates. Error bars represent standard error of mean (SEM). Statistical significance was determined by two-way ANOVA followed by H-Šídák’s multiple comparisons (****p < 0.0001). a, b are created with BioRender.com.