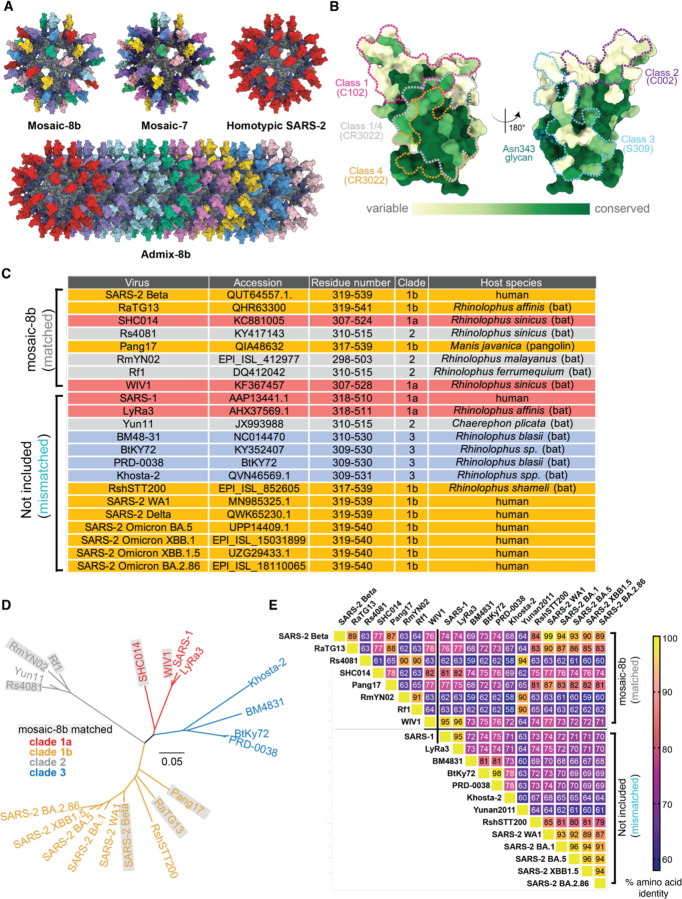
Figure 1. RBDs used to make nanoparticles and for assays, related to Figure S1, Table S1.
(A) Models of mosaic-8b, mosaic-7 (mosaic-8b without SARS-2 RBD), homotypic SARS-2, and admix-8b RBD-nanoparticles constructed using coordinates of an RBD (PDB 7BZ5), SpyCatcher (PDB 4MLI), and i3-01 nanoparticle (PDB 7B3Y).
(B) Sequence conservation determined using the ConSurf Database95 of the 16 sarbecovirus RBDs used to make nanoparticles and/or for assays shown on two views of an RBD surface (PDB 7BZ5). Class 1, 2, 3, 4, and 1/4 epitopes are outlined in different colors using information from Fab-RBD or Fab-spike trimer structures (C102, PDB 7K8M; C002, PDB 7K8T; S309, PDB 7JX3; CR3022, PDB 7LOP; and C118, PDB 7RKV).
(C) List of sarbecoviruses from which the RBDs in mosaic-8b and admix-8b were included (matched) or not included (mismatched). Clades were defined as described.96
(D) Phylogenetic tree of selected sarbecoviruses calculated using a Jukes-Cantor generic distance model using Geneious Prime® 2023.1.2 based on amino acid sequences of RBDs aligned using Clustal Omega.97 Viruses with RBDs included in mosaic-8b are highlighted in gray rectangles.
(E) Amino acid sequence identity matrix of RBDs based on alignments using Clustal Omega.97