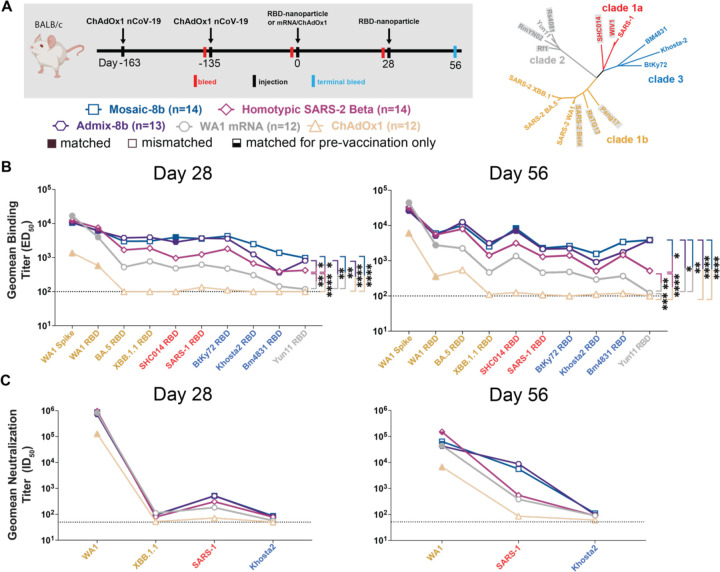
Figure 4. Mosaic-8b immunizations in ChAdOx1-vaccinated mice elicit cross-reactive Ab responses, related to Figure S4.
Geometric means of ED50 or ID50 values for all animals in each cohort are indicated by symbols connected by thick colored lines. Mean titers against indicated viral antigens or pseudoviruses were compared pairwise across immunization cohorts by Tukey’s multiple comparison test with the Geisser-Greenhouse correction (as calculated by GraphPad Prism). Significant differences between cohorts linked by vertical lines are indicated by asterisks: p<0.05 = *, p<0.01 = **, p<0.001 = ***, p<0.0001 = ****.
(A) Left: Schematic of vaccination/immunization regimen. Mice were vaccinated at the indicated days prior to RBD-nanoparticle prime and boost immunizations at days 0 and 28 or mRNA-LNP or ChAdOx1 prime immunizations at day 0. Right: Phylogenetic tree of selected sarbecoviruses calculated using a Jukes-Cantor generic distance model using Geneious Prime® 2023.1.2 based on amino acid sequences of RBDs aligned using Clustal Omega.97 RBDs included in mosaic-8b are highlighted in gray rectangles.
(B) Geometric mean ELISA binding titers at the indicated days after immunization with mosaic-8b, admix-8b, homotypic SARS-2, WA1 mRNA-LNP, or ChAdOx1 against indicated viral antigens.
(C) Geometric mean neutralization titers at the indicated weeks after immunization with mosaic-8b, admix-8b, homotypic SARS-2, WA1 mRNA-LNP, or ChAdOx1 against indicated sarbecovirus pseudoviruses.