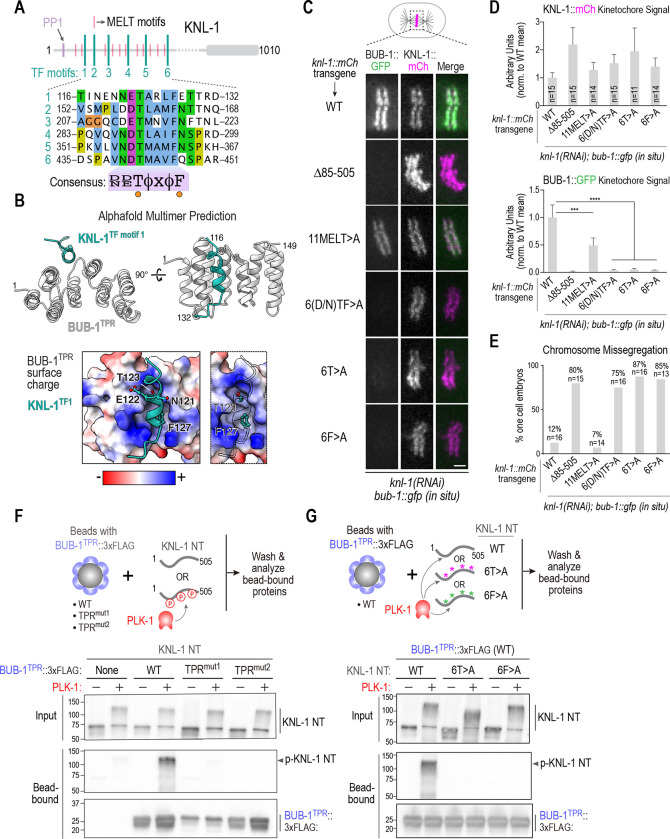
Figure 3. Definition of the BUB-1 TPR – KNL-1 interface that targets BUB-1 to kinetochores.
(A) Schematic of KNL-1 highlighting 11 MELT motifs, 6 TF motifs and protein phosphatase 1 (PP1) docking site. Sequences of the 6 TF motifs are shown below along with a consensus. (B) (top) AlphaFold multimer prediction of the BUB-1 TPR interface with TF motif 1. Side and en face views are shown. Predictions for all 6 TF motifs are shown in Fig. S2. (bottom) TF motif 1 shown docked onto a surface charge model of the BUB-1 TPR. Panel on the right highlights the T and F residues; the insertion of the F into a hydrophobic pocket is evident in this view. (C) (left) Images of aligned chromosomes in one-cell embryos from strains with the indicated knl-1::mCh transgenes and in situ GFP-tagged BUB-1; endogenous KNL-1 was depleted in all conditions. Scale bar, 2 μm. (D) Quantification of KNL-1::mCh and BUB-1::GFP kinetochore signals for the indicated conditions. n is the number of embryos imaged. Error bars are the 95% confidence interval. (E) Quantification of chromosome missegregation in one-cell embryos for the indicated conditions. n is the number of embryos imaged. (F) (top) Schematic of biochemical assay used to assess BUB-1 TPR interaction with the KNL-1 N-terminus, either with or without PLK-1 phosphorylation. The BUB-1 TPR variants were expressed in suspension human cells and concentrated on beads prior to mixing with unphosphorylated or PLK-1 phosphorylated bacterially expressed and purified KNL-1 N-terminus (NT). (bottom) Immunoblots of the KNL-1 input and bead-bound KNL-1 and BUB-1TPR variants. (G) Similar biochemical analysis as in (F), except that BUB-1TPR was WT in all conditions and three versions of recombinant KNL-1 NT (WT, 6T>A, 6F>A) were tested with and without PLK-1 phosphorylation. Data shown in (F) and (G) is representative of two independent experiments. All p values were calculated by unpaired t tests; *** = p<0.001,**** = p<0.0001.