Fig 1.
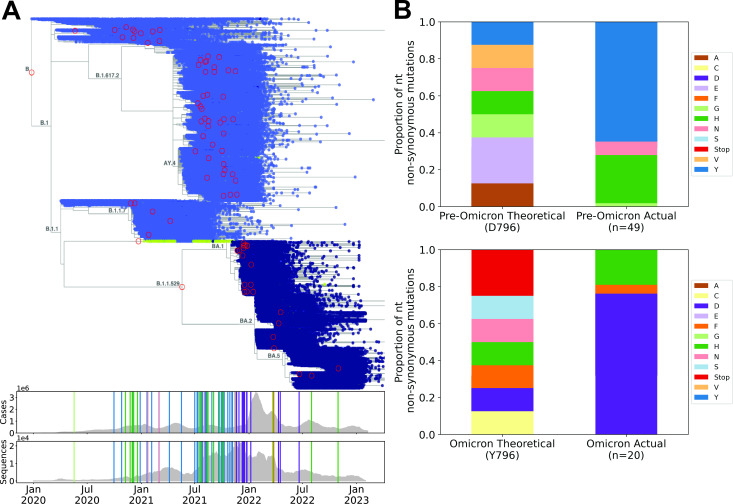
Phylogenetic analysis reveals that the D796Y and D796H substitutions occurred recurrently prior to the Omicron wave. (A) Upper: time-resolved phylogenetic tree including 6,329,365 SARS-CoV-2 sequences (Taxonium.org). The color of the points indicates the amino acid residue at position 796 of the SARS-CoV-2 Spike: light-blue, D; green, H; dark-blue, Y. Red circles highlight nodes with mutations at 796 that were found in ≥10 descendants (a threshold set to minimize the impact of technical artifacts). Branch labels indicate PANGO lineage (15). Lower: plots showing daily worldwide SARS-CoV-2 cases and total sequences per day present in the phylogenetic tree over a 2-year timescale (x-axis aligned with the phylogenetic tree in the upper panel). Vertical lines indicate the positions of mutants highlighted with red circles in the phylogenetic tree, and colors show the mutation type (matching the bar graphs in B). (B) The proportion of theoretically possible (left) compared with observed (right) non-synonymous amino acid substitutions resulting from single-nucleotide mutations in the codon for D796 in pre-Omicron sequences (upper) and Y796 in post-Omicron sequences (lower).