Abstract
Bacteroides forsythus is a recently recognized human periodontopathogen associated with advanced, as well as recurrent, periodontitis. However, very little is known about the mechanism of pathogenesis of this organism. The present study was undertaken to identify the surface molecules of this bacterium that may play roles in its adherence to oral tissues or triggering of a host immune response(s). The gene (bspA) encoding a cell surface-associated protein of B. forsythus with an apparent molecular mass of 98 kDa was isolated by immunoscreening of a B. forsythus gene library constructed in a lambda ZAP II vector. The encoded 98-kDa protein (BspA) contains 14 complete repeats of 23 amino acid residues that show partial homology to leucine-rich repeat motifs. A recombinant protein containing the repeat region was expressed in Escherichia coli, purified, and utilized for antibody production, as well as in vitro binding studies. The purified recombinant protein bound strongly to fibronectin and fibrinogen in a dose-dependent manner and further inhibited the binding of B. forsythus cells to these extracellular matrix (ECM) components. In addition, adult patients with B. forsythus-associated periodontitis expressed specific antibodies against the BspA protein. We report here the cloning and expression of an immunogenic cell surface-associated protein (BspA) of B. forsythus and speculate that it mediates the binding of bacteria to ECM components and clotting factors (fibronectin and fibrinogen, respectively), which may be important in the colonization of the oral cavity by this bacterium and is also a target for the host immune response.
Bacteroides forsythus, a gram-negative, anaerobic, fusiform bacterium first described by Tanner et al. (20), has recently been recognized as one of the periodontopathogens associated with periodontitis. The inability to reliably cultivate this organism from mixed infections of the oral cavity has precluded investigations of its importance in disease. Immunofluorescence (9), DNA-based probes (11), and PCR techniques (5) have been recently developed to obtain more precise estimates of its distribution in health and disease. Recent clinical studies of Grossi et al. have shown that the occurrence of B. forsythus in subgingival flora was significantly associated with the severity of periodontitis (attachment and alveolar bone loss) in adults and older patients (3, 4). Identification of virulence factors of B. forsythus would therefore aid in the development of preventive strategies against periodontal disease. Ultrastructurally, B. forsythus demonstrates smooth cell surface features likely devoid of any extracellular structures, suggesting unique surface antigens (9). The organism produces both a trypsin-like protease (21) and a sialidase (10). In addition, a prtH gene encoding a trypsin-like protease of B. forsythus has recently been cloned (15). Moreover, interbacterial binding between B. forsythus and Porphyromonas gingivalis that appears to involve protein-protein interactions has been suggested to play a role in the establishment of periodontopathic plaque (23).
We focused our studies on identifying surface antigens of B. forsythus that may play roles in the adherence and colonization of oral surfaces. In this report, we describe the cloning and expression of a B. forsythus surface antigen with a leucine-rich repeat (LRR) motif that may be involved in binding to fibronectin (an extracellular matrix [ECM] component) and fibrinogen (a clotting factor).
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
B. forsythus ATCC 43037 was grown anaerobically (85% N2, 10% H2, 5% CO2) in brain heart infusion (Difco Laboratories, Detroit, Mich.) broth containing 5-μg/ml hemin, 0.5-μg/ml menadione, 0.001% N-acetylmuramic acid (Sigma Chemical Co., St. Louis, Mo.), and 5% fetal bovine serum (Gibco-BRL, Grand Island, N.Y.) with or without 1.5% agar. The Lambda ZAP II cloning system for constructing a B. forsythus gene bank and Escherichia coli XL1-BlueMRF′ were purchased from Stratagene Inc., La Jolla, Calif. pGEX expression vectors were purchased from Pharmacia Inc., Piscataway, N.J. E. coli cells were grown in Luria-Bertani (LB) broth or LB agar (1.5%). Ampicillin (100 μg/ml) was added to broth or agar plates when needed.
Preparation of anti-B. forsythus sera.
New Zealand White male rabbits (3 kg) were immunized subcutaneously with 109 live B. forsythus ATCC 43037 cells emulsified in a copolymer adjuvant (TiterMax; Cytrex Corp., Norcross, Ga.) At 2 and 3 weeks after the primary immunization, the animals were given subcutaneous boosters of the same dose of bacteria without the adjuvant. The animals were bled 2 weeks after the last booster injection.
Construction of the B. forsythus gene library and screening and for the cell surface antigen gene.
Ten micrograms of chromosomal DNA isolated from fresh cultures of B. forsythus ATCC 43037 by utilizing Qiagen’s genomic DNA isolation kit (Qiagen Tip 100; Qiagen Inc., Chatsworth, Calif.) was partially digested with ApoI (New England Biolabs), and DNA fragments ranging in size from 2 to 7 kb were gel purified (Gene Clean; Bio 101, Inc., La Jolla, Calif.) following electrophoresis on a 0.8% agarose gel. A B. forsythus clone bank was then generated by cloning the gel-purified fragments into the EcoRI site of a Lambda Zap II vector as described by the supplier (Stratagene). The clone bank was screened for immunoreactive positive clones by using an anti-B. forsythus antibody. Several immunoreactive plaques were purified, amplified, and subjected to in vivo excision with the helper phage ExAssist and E. coli SOLR as the host as described in the Stratagene Lambda ZAP II instruction manual. The recombinant plasmids were then purified with an Easy Pure plasmid isolation kit (Primm Labs, Cambridge, Mass.) and subjected to restriction enzyme mapping.
Analysis of gene products by SDS-PAGE and immunoblotting.
Rescued phagemids obtained following in vivo excision of the positive plaques were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis. Briefly, a bacterial colony was grown in 4 ml of LB-ampicillin medium until the optical density at 600 nm (OD600) reached 0.6. Two-milliliter cultures were transferred to a fresh tube, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM. Both uninduced and IPTG-induced cultures were further incubated for 2 h, and cells were harvested by brief centrifugation. Cell pellets were washed once with TBS (0.1 M Tris, 0.15 M NaCl, pH 7.3) and resuspended in 200 μl of TBS containing 1 mM phenylmethylsulfonyl fluoride and sonicated for 30 s to obtain whole-cell lysates. Whole-cell lysates were then subjected to SDS-PAGE by the method of Laemmli utilizing the Mini-Tall gel system (Hoefer Scientific Instruments, San Francisco, Calif.), and the gels were stained with Coomassie brilliant blue. For immunoblot analysis, proteins separated by SDS-PAGE were transferred to nitrocellulose membranes by using a semidry transfer system (Semi-Phor TE-77; Hoefer Scientific Instruments). The membranes were probed with anti-B. forsythus serum, followed by horseradish peroxidase (HRP)-coupled goat anti-rabbit serum, and the color was developed.
Nested deletions, DNA sequencing, and determination of ORFs.
Collections of deletion subclones of recombinant plasmid pBBf4 were constructed by using the unique restriction sites of the plasmid for unidirectional deletions with exonuclease III (ExoIII) and S1 nuclease. pBBf4 was digested with BstXI and BamHI prior to the addition of ExoIII. ExoIII digestions were carried out to obtain sequential deletions of 250 to 300 bp at each time point. Deletion clones were subjected to SDS-PAGE and immunoblot analysis. Clones with immunoreactive protein bands (full-length or deletion-containing forms) were subjected to DNA sequence analysis to localize the open reading frames (ORFs). Double-stranded plasmid constructs were sequenced at the Nucleic Acid Core Facility (Center for Advanced Molecular Biology and Immunology) of the State University of New York at Buffalo with a 373A sequencer (Applied Biosystems, Foster City, Calif.). T3 and T7 promoter primers were utilized to sequence both strands. DNA data were analyzed by using the University of Wisconsin’s Genetics Computer Group software in conjunction with the Gene Inspector DNA and protein analysis software (Textco Inc.).
Southern blotting and PCR analysis.
Southern blotting and PCR analysis were performed to determine if the gene had been cloned in its entirety. The chromosomal DNA and positive clone pBBf4 were digested with restriction enzymes HincII and StyI. These two enzymes cut within the cloned sequence and yield bands of 2.1 and 2.9 kb, respectively, when probed with the 2.1-kb internal fragment of the bspA gene. After electrophoresis on 1% agarose gels, the separated DNA fragments were transferred onto a Hybond membrane by the capillary transfer technique. DNA fragments were cross-linked to the membrane by a UV cross-linker (Stratagene) and probed with PCR-amplified fragments or restriction fragments of the cloned gene labeled with an Amersham Life Sciences nonradioactive labeling kit in accordance with the manufacturer’s recommendations.
PCR was performed by using an Expand High Fidelity PCR system (Boehringer Mannheim, Indianapolis, Ind.) in an Omn-E thermal cycler (Hybaid Ltd.) in accordance with the manufacturer’s recommendations. Amplimers for PCR were based on the nucleotide sequence of the cloned gene. Custom oligonucleotide primers were synthesized at the Center for Advanced Molecular Biology and Immunology nucleic acid facility (State University of New York at Buffalo).
pGEX expression and biotin labeling.
Expression of the recombinant protein was carried out with pGEX expression vectors (Pharmacia Inc., Piscataway, N.J.). Briefly, the internal HincII fragment of bspA encoding residues 17 to 724 (LRR region) of the BspA protein was cloned in frame with the glutathione S-transferase (GST) gene in the SmaI site of pGEX-2T. Protein expression was carried out in accordance with standard protocols (1), and the expressed fusion protein was bound to glutathione agarose beads (Sigma Chemical Co., St. Louis, Mo.) following lysis of the E. coli cells by sonication. The bead-bound fusion protein was treated with thrombin (0.1% [wt/wt]; Sigma Chemical Co.) for 2 h at room temperature, and the cleaved recombinant protein was eluted and analyzed by SDS-PAGE.
The recombinant protein was coupled to biotin by the reagent N-hydroxysuccinimidobiotin (Pierce Chemical Co., Rockford, Ill.). Briefly, purified protein (2 mg/ml) was dialyzed against 50 mM sodium bicarbonate buffer (pH 8.5) and reacted for 30 min at room temperature with 25 μl of the N-hydroxysuccinimidobiotin reagent (1 mg dissolved in dimethyl sulfoxide). Unreacted reagent was removed by dialysis against 50 mM Tris-Cl (pH 7.2) buffer, and the biotin-labeled protein was stored at −20°C.
Antibody production.
SDS-PAGE-separated and Coomassie blue R250-stained protein bands were gel eluted by utilizing a Gel Eluter (Isco Inc., Lincoln, Nebr.). Eluted protein (100 to 150 μg) was emulsified in a copolymer adjuvant (TiterMax) and injected subcutaneously at 2-week intervals into New Zealand White male rabbits (3 kg). Two weeks later, blood was withdrawn and antiserum was tested by immunoblotting.
Binding and binding inhibition assays.
Microtiter plate wells were coated with protein ligands (fibrinogen and fibronectin) by incubating each well with 2 μg of protein dissolved in 0.1 M sodium bicarbonate buffer, pH 9.6 (100 μl/well), at room temperature for 2 h. Unoccupied sites were blocked with 200 μl of 0.5% fat-free bovine serum albumin (BSA; Sigma Chemical Co.) in phosphate-buffered saline (PBS) for 1 h at room temperature. Wells were then incubated with 50 μl of B. forsythus cells prewashed with PBS containing 0.02% azide and brought to an OD600 of 0.125. For the binding inhibition assays, purified rBsp70 (a 70-kDa recombinant protein obtained by thrombin cleavage of the GST-Bsp fusion protein) was added at increasing concentrations in 50 μl to wells containing B. forsythus cells. Control wells contained B. forsythus cells and PBS only. The reaction mixture was incubated for 1 h at room temperature and then washed three times with PBS containing 0.05% Tween 20 (PBST). Bound cells were fixed with a 2% formaldehyde solution, washed with PBST, and incubated for 1 h at room temperature with an anti-B. forsythus antibody (1:5,000 dilution in PBS-BSA). The wells were then washed with PBST and incubated with goat anti-rabbit immunoglobulin G (IgG) coupled to HRP (1:1,000 in PBS-BSA; Bio-Rad, Inc., Richmond, Calif.). After the wells had been washed with PBST, color was developed with the substrate reagent TMB (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.). Cell binding was quantified by measuring OD595 with a Bio-Rad microplate reader. Percent binding inhibition was calculated as [(OD595 of control well − OD595 of rBsp70 well)/OD595 of control]) × 100.
Overlay assays.
Two methods were employed to evaluate binding to proteins. In the first method, proteins (fibronectin, fibrinogen, type I collagen, and BSA) were blotted onto a nitrocellulose membrane by a slot blot apparatus (Pharmacia Inc.) and the unoccupied sites were blocked by incubating the membrane at room temperature for 1 h with 0.5% BSA. In the second method, serial dilutions of fibrinogen or fibronectin were mixed with a 50-μg/ml BSA solution, 100 μl was blotted onto a membrane, and the membranes were blocked with BSA as described above. The membranes were then incubated with biotin-labeled recombinant protein for 1 h at room temperature and washed with PBST. The bound proteins were detected by incubating the membrane with peroxidase-labeled streptavidin, and color was developed with the membrane substrate reagent TMB (Kirkegaard & Perry Laboratories, Inc.).
Immunofluorescence analysis.
Immunofluorescence analysis was performed on nonpermeabilized, heat-fixed B. forsythus cells. Briefly, a loopful of actively growing B. forsythus cells obtained from agar plates was resuspended in 2 ml of Ringer’s solution containing 2% formalin and a drop was allowed to dry on a microscope slide. Bacterial cells were then briefly heat fixed, incubated with anti-rBsp70 or preimmune rabbit serum, washed, and incubated with goat anti-rabbit IgG coupled with fluorescein isothiocyanate. Photographs were obtained by using a Nikon epifluorescence microscope and the Bio-Rad MRC-1024 (Bio-Rad) three-channel laser scanning confocal imaging system equipped with a krypton-argon laser.
Nucleotide sequence accession number.
The sequence of the bspA gene has been deposited in the GenBank database under accession no. AF054892.
RESULTS
Cloning and expression of the B. forsythus surface antigen.
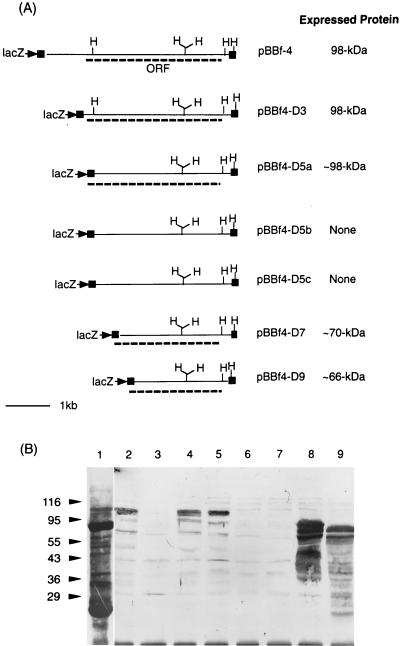
A gene bank of B. forsythus was constructed by cloning partially ApoI digested 2- to 7-kb chromosomal DNA fragments into the EcoRI site of the Lambda ZAP II vector. Approximately 30,000 plaques were replica blotted onto nitrocellulose filters and probed with polyclonal B. forsythus antisera preadsorbed with E. coli XL1 BlueMRF′ extract. Of five positive clones obtained, one strongly reactive phage clone was plaque purified, amplified, and rescued as a recombinant phagemid (pBluescriptSK derivative; Stratagene) by in vivo excision with a helper phage. The rescued phagemid was designated pBBf4. Samples of the phage lysates and E. coli cells harboring phagemid pBBf4 were subjected to SDS-PAGE and immunoblotting to detect expressed proteins. Plasmid pBBf4 carrying a 4.2-kb insert was utilized for restriction enzyme mapping and construction of deletion mutants.
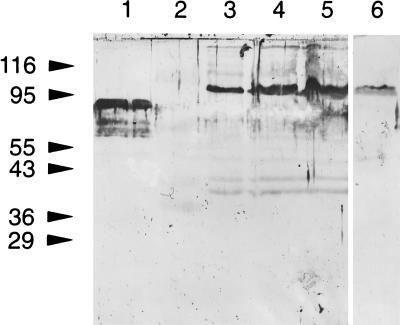
The banding patterns of SDS-PAGE-separated and Coomassie blue-stained proteins of the positive E. coli clone (extracts containing pBBf4) did not show any noticeable differences from that of controls (E. coli extracts containing pBluescriptSK; data not shown). However, immunoblot analysis of the same proteins by using anti-B. forsythus serum detected a protein band at 98 kDa for the positive clone (Fig. 1B, lane 2). No immunoreactive bands were detected in control lanes. As expected, in addition to a similar-size (98-kDa) band, multiple bands reacted in a whole-cell extract of B. forsythus with anti-B. forsythus whole-cell serum (Fig. 1B, lane 1).
FIG. 1.
(a) Restriction maps of positive clone pBBf4 and ExoIII-S1 nuclease deletion clones. The cloned fragment in positive clone pBBf4 is 4.2 kbp. The portions of the plasmid sequence flanking the cloned insert are shown as filled closed boxes. The ORF for the bspA gene in pBBf4 and the lacZ-bspA fusions in deletion clones are shown as dotted lines. H, HincII. (b) Immunoblot analysis of the positive clone and the ExoIII deletions. E. coli extracts harboring positive clone pBBf4 and the 5′ deletions of the insert were transferred onto nitrocellulose membranes after separation by SDS-PAGE. Membranes were probed with rabbit anti-B. forsythus serum followed by goat anti-rabbit IgG coupled to HRP. Membranes were developed with the color developing reagent TMB. Lanes: 1, total B. forsythus cell extract; 2, pBBf4 (positive clone); 3, pBluescriptSK (negative control); 4, pBBf4-D3; 5, pBBf4-D5a; 6, pBBf4-5b; 7, pBBf4-D5c; 8, pBBf4-D7; 9, pBBf4-D9. The positions of molecular size markers (sizes are in kilodaltons) are shown on the left.
Expression of the 98-kDa protein was inducible following addition of IPTG to the growing cultures of positive clones (E. coli containing pBBf4). However, cultures incubated in the absence of IPTG showed little or no expression (data not shown). These results indicated either that the cloned bspA gene is devoid of its own promoter sequences or that such sequences are not recognized in E. coli.
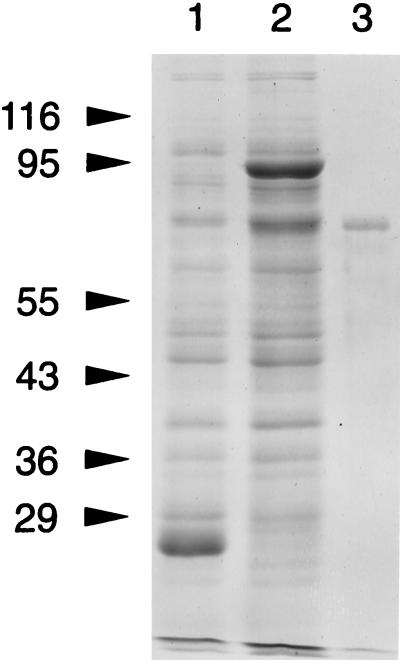
The expression of a 2.1-kb HincII fragment in pGEX-2T resulted in the expression of a GST-fusion protein which, on thrombin cleavage, resulted in a recombinant protein (rBspA, residues 17 to 724) with an apparent molecular mass of ∼70 kDa as determined by SDS-PAGE (Fig. 2). The sequence of the 10 amino-terminal residues of the rBspA protein determined by protein sequencing (ProSeq Sequencing Service, Salem, Mass.) was found to be identical to the predicted amino acid sequence of the thrombin-cleaved product (NH2-GlySerProThrThrLeuGlyAlaThrAla...COOH). The amino-terminal residues, GlySerPro, were encoded by the vector sequence.
FIG. 2.
SDS-PAGE analysis of recombinant protein rBsp70. Lanes: 1, E. coli cells harboring pGEX-2T; 2, E. coli cells harboring pGEX-2.1 HincII; 3, purified recombinant protein rBsP70. The values on the left are molecular sizes in kilodaltons.
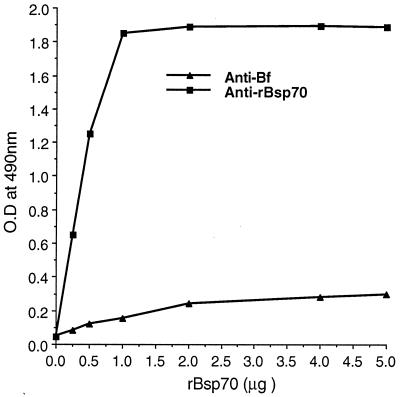
Additionally, high anti-rBsp70 antibody titers were detected in the serum of rabbits immunized with reduced and denatured SDS-PAGE-purified protein by enzyme-linked immunosorbent assay (ELISA), whereas rBsp70 did not react with whole-cell anti-B. forsythus rabbit serum. As shown in Fig. 3, rBsp70 used to coat microtiter plate wells reacted strongly with anti-rBsp70 compared to anti-B. forsythus antibody at a similar dilution (1:5,000). The anti-B. forsythus antibodies probably recognize linear and/or conformational epitopes outside of the region present in rBsp70 (amino acid residues 17 to 724).
FIG. 3.
Reactivity of rBsp70 with anti-B. forsythus (Bf) and anti-rBsp70 antibodies as determined by ELISA. Microtiter plate wells coated with purified protein rBsp70 were incubated with 1:5,000-diluted rabbit anti-B. forsythus or rabbit anti-rBsp70 serum and then incubated with goat anti-rabbit IgG coupled to HRP, and color was developed with the substrate reagent TMB. Absorbance was plotted against rBsp70 for each of the two antibodies.
Nested deletions, DNA sequencing, and identification of ORFs.
In order to identify the ORF for the 98-kDa protein and sequence its gene, parts of pBBf4 were unidirectionally deleted by using the ExoIII-S1 nuclease procedure. The plasmid subclones with deletions were analyzed for expression of the 98-kDa protein by immunoblot analysis. Deletion-containing subclones that express immunoreactive protein bands were characterized by restriction mapping and partial DNA sequencing with a T3 primer at the junction of the lacZ gene and the deletion-containing insert. Restriction maps of inserts with unidirectional deletions excised from the plasmid subclones with PvuII (this enzyme cuts outside the insert DNA in pBluescriptSK) and the corresponding ORFs are summarized in Fig. 1A. A 98-kDa immunoreactive band was observed in plasmid subclones pBBf4-D3 and pBBf4-D5a, whereas smaller immunoreactive bands of ∼70 and ∼66 kDa were observed in deletion subclones pBBf4-D7 and pBBf4-D9, respectively (Fig. 1B). The 98-kDa ORF was therefore localized on the deletion fragment in subclone pBBf4-D3. In addition, DNA sequencing confirmed that the lacZ gene was in frame with the 98-kDa ORF with deletions at the 5′ end in subclones pBBf4-D5a, pBBf4-D7, and pBBf4-D9, respectively, thereby expressing N-terminally truncated polypeptides of the 98-kDa protein. Deletion clones pBBf4-D5b and pBBf4-5c, which did not express immunoreactive bands, were not in frame with the lacZ gene.
Sequence analysis of the cloned 98-kDa protein.
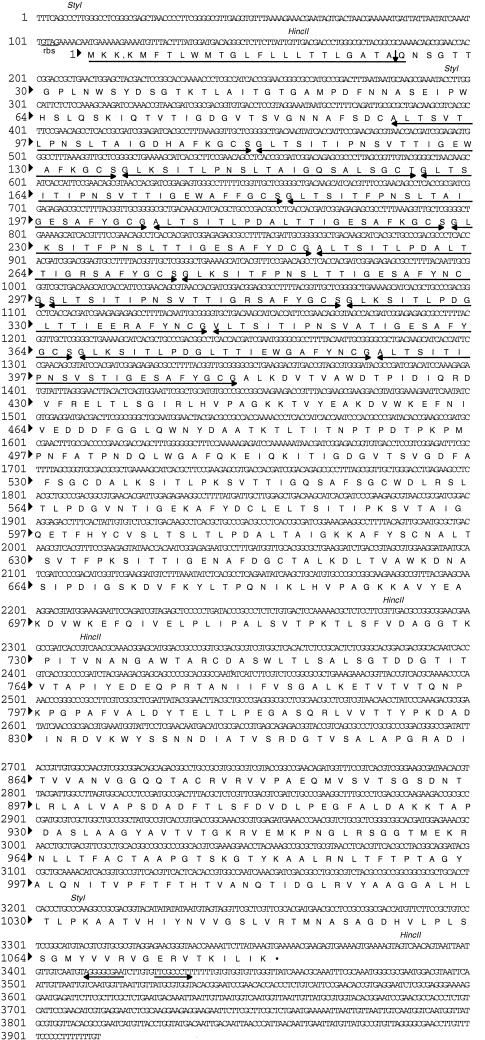
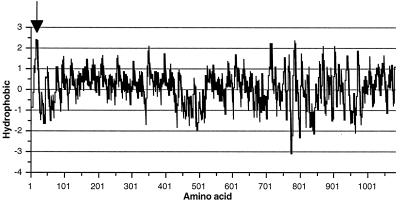
As mentioned above, a single long ORF was identified on deletion subclone pBBf4-D3. DNA sequencing results identified an ORF with a potential translational start site at base position 112, which is 6 bp downstream from a putative ribosomal binding site (17), GTAG (Fig. 4). The actual sequences of the B. forsythus ribosome-binding sites have not been determined. Due to the highly repetitive nature of the DNA sequence between nucleotide residues 385 and 1346 of the gene, causing multiple priming of the sequencing primers, deletions were made in the insert DNA in both directions and the DNA was sequenced with vector primers. The ORF would encode a 1,081-amino-acid protein with a deduced molecular mass of 113,921 Da and a pI of 5.4. There is a sequence resembling a rho-independent transcription termination sequence from nucleotide 3411 to nucleotide 3436 that contains an inverted repeated sequence. A stretch rich in T residues much like typical E. coli terminators immediately follows the inverted repeat structure (Fig. 4). The gene was named bspA (B. forsythus surface protein A), and the encoded protein was termed BspA. Alternatively, initiation at base position 265 would yield a slightly smaller protein with 1,030 amino acids, a calculated molecular mass of 108,532 Da, and a pI of 5.17. The molecular masses deduced from both of the ORFs were larger than the apparent molecular mass determined by SDS-PAGE analysis. This discrepancy is probably due to the anomalous migration of the protein on SDS-PAGE. A Kyte-and-Doolittle hydropathy plot (Fig. 5) of the deduced protein revealed a hydrophobic N terminus which has a low surface probability and exhibits features similar to those of signal peptides of E. coli (14). The N-terminal sequence of 23 amino acids has three charged lysines in the first six residues and is followed by a hydrophobic core with a potential signal peptidase site at A-24 to Q-25 (Fig. 4) which would satisfy the criteria of von Heijne (22). It is important to mention here that the suggested start codon (ATG, residues 113 to 115) and signal peptide sequence are speculative pending determination of the N-terminal amino acid sequence of the native protein, or at least that of the BspA protein encoded by positive clone pBBf4.
FIG. 4.
Nucleotide sequence and the deduced amino acid sequence of the BspA protein. The potential leader peptide sequence is underlined, and the possible cleavage site is shown by a downward-pointing arrow. The LRR region is underlined with double-head arrows. A putative ribosomal binding site (rbs) and two restriction sites (HincII and StyI) are shown. A possible terminator is indicated by the back-to-back arrows.
FIG. 5.
Kyte-Doolittle hydropathy plot of the deduced amino acid sequence encoded by the bspA gene of B. forsythus. Hydrophobic regions are above the line, and the hydrophobic index is indicated on the vertical axis. Numbers on the horizontal axis refer to positions in the ORF. A putative signal sequence is indicated by the arrow.
Structural analysis and identification of motifs.
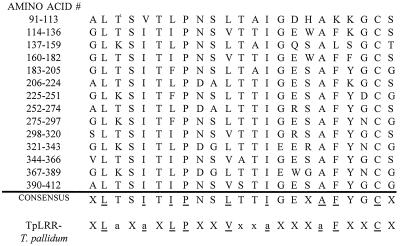
A BLAST search of the entire BspA sequence failed to identify any significant homologies to known proteins in the database at the National Center for Biotechnology Information, Bethesda, Md. In contrast, visual inspection of the deduced amino acid sequence and restriction of the searches to repeat regions of the protein showed significant homology with the TpLRR protein of Treponema pallidum (16). More-detailed analysis revealed that the periodicity was due to 14 tandemly arranged 23-amino-acid repeats followed by a 15th, incomplete repeat (Fig. 6). The consensus sequence for these repeats matched that for the T. pallidum TpLRR protein, an LRR protein. LRR motifs have been identified in a large number of eukaryotic and prokaryotic proteins with diverse functions and cellular localizations (8). However, the LRR motif of BspA and TpLRR differed from the LRR superfamily motif, since the presence of a turn promoting proline at position 8 and a cysteine residue at position 22 differed from the turn promoting an asparagine, threonine, or cysteine residue at position 10 found in other known proteins of the LRR superfamily.
FIG. 6.
Alignment of the 23-aminno-acid repeats of the BspA protein and the T. pallidum LRR protein (TpLRR). The amino acid positions of the BspA protein are indicated on the left. The one-letter amino acid code is used. An amino acid is included in the consensus if it is present at that position in more than half of the repeats. Amino acid positions with identical or a similar amino acid substitutions between the BspA consensus and the TpLRR consensus are underlined. X, any amino acid; a, alanine, valine, leucine, isoleucine, phenylalanine, tyrosine, or methionine; N, asparagine, cysteine, or threonine.
Binding and binding inhibition assays.
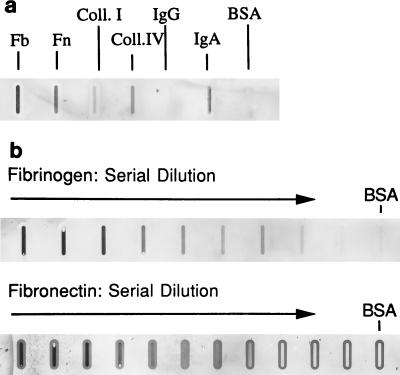
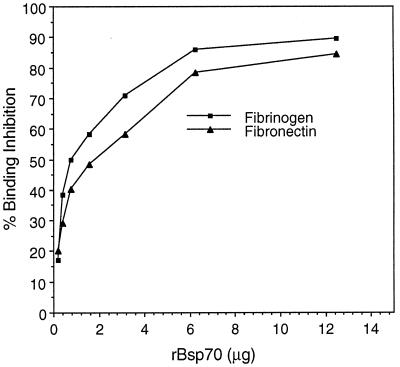
The presence of a repeat motif in BspA suggested that this protein may exhibit adhesive activity. The overlay assay results showed that rBsp70 bound fibrinogen and fibronectin in a dose-dependent manner. It did not bind type I collagen or BSA (Fig. 7). Moreover, visual qualitative analysis showed that rBsp70 bound more strongly to fibrinogen than to fibronectin. These results were confirmed by a quantitative binding ELISA (data not shown). In addition, rBsp70 inhibited the binding of B. forsythus cells to fibrinogen and fibronectin in a dose-dependent manner (Fig. 8). A maximum of 85 to 90% inhibition of B. forsythus binding to ECM components was achieved at saturating concentrations of rBsp70. The above results, taken together, suggest that the BspA protein is involved in the colonization of the oral cavity by B. forsythus via binding to ECM components.
FIG. 7.
Overlay assay. (a) Binding of rBsp70 to surface-bound protein ligands. One micogram of each of the proteins (from the left: 1, fibronectin (Fb); 2, fibrinogen (Fn); 3, type I collagen (Coll. I); 4, BSA; 5, saline blank) was blotted onto a nitrocellulose membrane. The membrane was probed with biotin-labeled rBsp70, followed by a streptavidin-peroxidase conjugate, and color was developed with the substrate TMB. The membranes in panel b were blotted with twofold serial dilutions of 4 μg of fibrinogen (or fibronectin) mixed with 4 μg of BSA. The membranes were then probed with biotin-labeled rBsp70, followed by a streptavidin-peroxidase conjugate, and color was developed with TMB as described above.
FIG. 8.
Binding inhibition assay. Inhibition of B. forsythus binding to surface-coated matrix components by rBsp70. Microtiter plate wells coated with matrix components were incubated with B. forsythus cells in the presence of increasing concentrations of the rBsp70 protein. Bound cells were detected by an anti-B. forsythus antibody, followed by a second antibody coupled to peroxidase, and color development with TMB. Percent inhibition of B. forsythus cell binding to each matrix component was plotted against the rBsp70 concentration. The results shown are means of duplicate samples that are representative of several experiments.
Immunoblot analysis of B. forsythus cells.
Total cell extracts, spent medium, and cell envelopes of B. forsythus were transferred onto nitrocellular membranes after separation by SDS-PAGE. The envelope fraction was prepared by a previously described procedure for gram-negative bacteria (7). Membranes were probed with a rabbit anti-rBsp70 antibody and goat anti-rabbit IgG coupled to peroxidase and developed with the substrate reagent TMB to detect the bound antibody. A 98-kDa band reacted strongly in lanes containing total cell extracts or the envelope fraction with the anti-rBsp70 antibody (Fig. 9). We also noticed a weakly reacting band with a similar size (98 kDa) in the medium of B. forsythus cell cultures. These results confirmed that the positive clone expresses a full-length protein similar in size to the protein associated with the cell envelope of B. forsythus.
FIG. 9.
Immunoblot analysis of the B. forsythus membrane fraction. SDS-PAGE-separated proteins were transferred onto nitrocellulose membranes and probed with a rabbit anti-rBsp70 antibody and goat anti-rabbit IgG coupled to HRP. The enzyme reaction was developed with the membrane substrate TMB. Lanes: 1, rBsp70; 2, E. coli cell extract containing pBluescrioptSK− (used as negative control); 3, E. coli extract harboring pBBf4 (positive clone); 4, B. forsythus membrane fraction; 5, B. forsythus whole-cell extract; 6, culture medium of actively growing B. forsythus cells. The values on the left are molecular sizes in kilodaltons.
Immunofluorescence analysis.
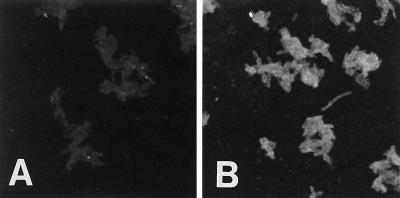
To detect the BspA protein on the B. forsythus cell surface, we carried out immunofluorescence microscopy. An anti-rBsp70 antibody was utilized for labeling of nonpermeabilized B. forsythus cells. Cell surface-associated fluorescence was detected under a fluorescence microscope on B. forsythus cells reacted with the anti-rBsp70 antibody (Fig. 10). Cells reacted with the preimmune serum used as a negative control did not show any fluorescence.
FIG. 10.
Immunofluorescence of B. forsythus cells. B. forsythus cells were incubated with rabbit anti-rBsp-70 serum (B) or preimmune rabbit serum (A), followed by goat anti-rabbit IgG coupled to fluorescein isothiocyanate. Cells reacted with the anti-rBsp70 antibody show positive fluorescence. Original magnification, ×425.
Human antibody response.
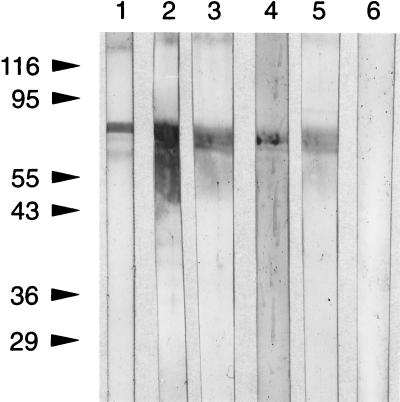
We examined the sera of periodontitis patients with oral flora that tested positive for the presence of B. forsythus and high titers of antibodies against whole cells to determine if a specific antibody response against BspA is generated. Briefly, the thrombin-cleaved product of the GST fusion protein separated by SDS-PAGE was transferred onto nitrocellulose membranes. Following transfer, the membrane was blocked with BSA, individual strips were cut out, and each was incubated with individual patient or control human serum (1:250 dilution in PBS-BSA). This was followed by washing and detection of the bound antibody with peroxidase-labeled goat anti-human IgG and color development with the membrane substrate TMB. The results showed that the rBsp70 protein reacted with the sera of all five of the periodontitis patients examined by immunoblot analysis (Fig. 11). Human serum from a healthy individual whose oral flora was devoid of B. forsythus did not react with rBsp70. These results indicate that the BspA protein may be an immunodominant protein in humans and a target for a host immune response.
FIG. 11.
Human antibody response. The thrombin-cleaved product (rBsp70) of the GST fusion protein was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Following blocking with BSA, individual membrane strips were cut out and each was incubated with individual patient and control human sera (1:250 dilution in PBS-BSA). The membrane was then developed with HRP-coupled with goat anti-human IgG and the membrane substrate reagent TBM. Membrane strips 1 to 5 were incubated with individual patient sera positive for B. forsythus, whereas strip 6 was incubated with the serum of a healthy individual (no periodontal disease). The values on the left are molecular sizes in kilodaltons.
DISCUSSION
We report here the cloning and expression of a B. forsythus gene, bspA, expressing a protein (BspA) which may be involved in binding to fibronectin (an ECM component) and fibrinogen (a clotting factor). BspA is a 98-kDa protein based on the results of immunoblotting of the total cell extracts of the positive E. coli clone and B. forsythus cells. The protein appears to be acidic in nature with a pI of 5.4. The deduced amino acid sequence of the gene encoding BspA showed 14 complete repeats of 23 amino acid residues (consensus: XLTSITIPNSLTTIGEXAFYGCX) and a 15th, incomplete repeat. The BLAST search of the repeat region at the National Center for Biotechnology Information showed homology with the T. pallidum LRR protein (TpLRR). The LRRs are proposed to be involved in protein-protein or lipid-protein interactions and may act as adhesins (8).
To determine if BspA containing repeat sequences which may be involved in protein-protein interactions plays any role in adherence, we carried out in vitro studies involving direct binding to fibronectin and fibrinogen. For this purpose, a portion of BspA (residues 17 to 724) containing the repeat region was expressed in E. coli by utilizing the pGEX expression system and the 70-kDa recombinant protein (rBsp70) was purified. In vitro studies showed that rBsp70 bound fibronectin and fibrinogen and inhibited the binding of B. forsythus to these components in a dose-dependent manner. Since the inhibition of binding of B. forsythus to fibronectin or fibrinogen by rBsp70 reached saturation at 80 to 85%, it is possible that the region downstream from the repeat sequence corresponding to amino acid residues 725 to 1081 also plays a role in binding. Alternatively, other cell surface components may be involved. Our preliminary studies showed that B. forsythus whole cells bound to fibronectin and fibrinogen in a dose-dependent and saturable manner much more strongly than to BSA (binding to fibronectin and fibrinogen was 8- to 10-fold higher than that to BSA). Moreover, we found that free arginine could inhibit the binding of B. forsythus to fibronectin or fibrinogen in a dose-dependent manner. However, free sugars were ineffective as inhibitors (data not shown). These results indicated that protein-protein interactions between cell surface components and the surface-bound matrix component(s) are likely involved in bacterial adherence. Studies by others (18) have also shown that Bacteroides spp. bind fibronectin via protein-protein interactions. Taken together, our results indicate that B. forsythus binding to the matrix components fibrinogen and fibronectin is mediated, at least in part, by BspA.
In addition to its role in adherence, the BspA protein may also be involved in host immune response modulation. The positive reaction of rBsp70 with antiserum from periodontitis patients testing positive for B. forsythus suggests that BspA is strongly immunogenic. It is also of interest that the proposed cytokine-producing domain (consensus: XLTX..XLT) proposed by Hamada et al. (12) for P. gingivalis fimbriae is also present in the BspA protein at residues 606 to 706. It has been proposed that this domain, which is also found in other bacteria (2, 13), induces the production of proinflammatory cytokines such as interleukin-1β (IL-1β), IL-6, IL-8, and tumor necrosis factor alpha in human peripheral blood monocyte-macrophage cultures. Our future studies will focus on determining if, in addition to its role in adherence, BspA could also play a direct role in inflammation. The fimbrial subunit protein (fimbrillin) of P. gingivalis has been shown to play a multifunctional role in the adherence of bacteria in the oral cavity and in the immunological stimulation of the host (6). In addition, our studies showed that BspA did not have any protease activity, since the total cell extracts of the positive clone had no activity detectable by zymography (casein gels) in the 98-kDa region (data not shown). In addition, we did not observe any protease activity of this size in total cell extracts of B. forsythus. A weak hemagglutinating activity was, however, detected in the purified rBsp70 protein. The significance of this activity is not known, but it is likely due to cross-linking of sheep blood cells via protein-protein interactions.
B. forsythus is frequently isolated together with P. gingivalis, which indicates an ecological relationship between the two species. Interbacterial binding between P. gingivalis and B. forsythus has recently been shown (23). In addition, B. forsythus was found to be highly virulent and invasive in combination with P. gingivalis in a rabbit model involving abscess formation (19). In light of the above findings, it is tempting to propose a role for the ECM binding properties of BspA in the colonization and virulence of B. forsythus. P. gingivalis degradation of host tissue could cause surface exposure of the ECM and its components for further colonization by B. forsythus, thereby enhancing the disease. The role of BspA in the pathogenic process could be examined by constructing BspA-negative mutants of B. forsythus and subsequent coinoculation of B. forsythus mutants with P. gingivalis in the rabbit abscess model. Development of a gene knockout system for B. forsythus is under investigation in our laboratory.
ACKNOWLEDGMENTS
We thank Robert Summers of the Imaging Facility in the Department of Microbiology, SUNY, Buffalo, for his expert assistance with confocal immunofluorescence microscopy and Linda Holben for her expert assistance with the binding studies.
This work was supported by PHS grants DE04898 and DE12320-01.
REFERENCES
- 1.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 2.Bakker D, Willemsen P T J, Simons L H, van Zijderveld F G, de Graff F K. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol Microbiol. 1992;6:247–255. doi: 10.1111/j.1365-2958.1992.tb02006.x. [DOI] [PubMed] [Google Scholar]
- 3.Grossi S G, Genco R J, Machtei E E, Ho A W, Koch G, Dunford R G, Zambon J, Hausmann E. Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J Periodontol. 1995;66:23–29. doi: 10.1902/jop.1995.66.1.23. [DOI] [PubMed] [Google Scholar]
- 4.Grossi S G, Zambon J J, Ho A W, Koch G, Dunford R G, Machtei E E, Norderyd O M, Genco R J. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 5.Guillot E, Mouton C. A PCR-DNA based assay specific for Bacteroides forsythus. Mol Cell Probes. 1996;10:413–421. doi: 10.1006/mcpr.1996.0057. [DOI] [PubMed] [Google Scholar]
- 6.Hamada S, Fujiwara T, Morishima S, Takahashi I, Nakagawa I, Kimura S, Ogawa T. Molecular and immunological characterization of the fimbriae of Porphyromonas gingivalis. Microbiol Immunol. 1994;38:921–930. doi: 10.1111/j.1348-0421.1994.tb02148.x. [DOI] [PubMed] [Google Scholar]
- 7.Joe A, Yamamoto A, McBride B C. Characterization of recombinant and native forms of a cell surface antigen of Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1993;61:3294–3303. doi: 10.1128/iai.61.8.3294-3303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 9.Lai C-H, Listgarten M A, Shirikawa M, Slots J. Bacteroides forsythus in adult gingivitis and periodontitis. Oral Microbiol Immunol. 1987;2:153–157. doi: 10.1111/j.1399-302x.1987.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 10.Moncla B J, Braham P, Hillier S L. Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacterial. J Clin Microbiol. 1990;28:422–425. doi: 10.1128/jcm.28.3.422-425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moncla B J, Motley S T, Braham P, Ewing L, Adams T H, Vermeulen N M J. Use of synthetic oligonucleotide DNA probes for identification and direct detection of Bacteroides forsythus in plaque samples. J Clin Microbiol. 1991;29:2158–2162. doi: 10.1128/jcm.29.10.2158-2162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalis fimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–242. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 13.Paranchych W, Sastry P A, Volpel K, Loh B A, Speert D P. Finbriae (pili): molecular basis of Pseudomonas aeruginosa adherence. Clin Investig Med. 1986;9:113–118. [PubMed] [Google Scholar]
- 14.Rusch S L, Kendall D A. Transport of an export-defective protein by a highly hydrophobic signal peptide. J Biol Chem. 1994;269:1243–1248. [PubMed] [Google Scholar]
- 15.Saito T, Ishihara K, Kato T, Okuda K. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect Immun. 1997;65:4888–4891. doi: 10.1128/iai.65.11.4888-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevchenko D V, Akins D R, Robinson E, Li M, Popova T G, Cox D L, Radolf J D. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum the syphilis spirochete. J Bacteriol. 1997;179:3188–3195. doi: 10.1128/jb.179.10.3188-3195.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shine J, Dalgarno L. The 3′ terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosomal binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szoke I, Pascu C, Nagy E, Ljung A, Wadstrom T. Binding of extracellular matrix proteins to the surface of Bacteroides sp. Anaerobe. 1997;3:91–95. doi: 10.1006/anae.1997.0079. [DOI] [PubMed] [Google Scholar]
- 19.Takemoto T, Kurihara H, Dahlen G. Characterization of Bacteroides forsythus isolates. J Clin Microbiol. 1997;35:1378–1381. doi: 10.1128/jcm.35.6.1378-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner A C R, Listgarten M A, Ebersole J L, Strzempko M N. Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 1986;36:213–221. [Google Scholar]
- 21.Tanner A C R, Strzempko M N, Belsky C A, McKinley G A. API-Zym and API-ANADENT reactions of fastidious gram-negative species. J Clin Microbiol. 1985;22:333–335. doi: 10.1128/jcm.22.3.333-335.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Heijne G. Structural and thermodynamic aspects of the transfer of proteins into and across membranes. Curr Top Membr Transp. 1985;242:151–179. [Google Scholar]
- 23.Yao E S, Lamont R J, Leu S P, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. 1996;11:35–41. doi: 10.1111/j.1399-302x.1996.tb00334.x. [DOI] [PubMed] [Google Scholar]