Extended Data Fig. 1: Publicly available browser for sharing phasing data.
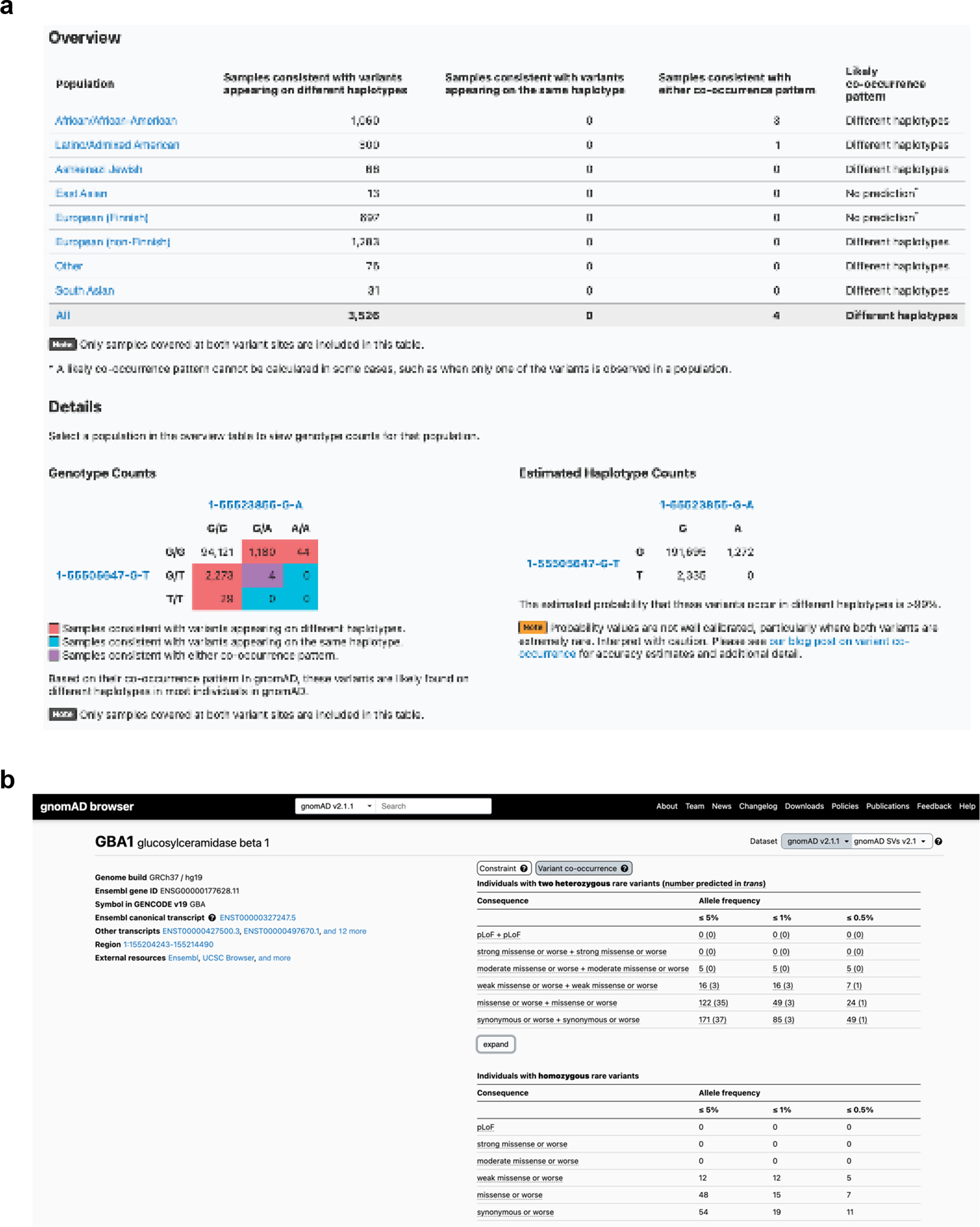
a, Sample gnomAD browser output for two variants (GRCh37 1–55505647-G-T and 1–55523855-G-A) in the gene PCSK9. On the top, a table subdivided by genetic ancestry group displays how many individuals in gnomAD v2 from that genetic ancestry are consistent with the two variants occurring on different haplotypes (trans), and how many individuals are consistent with their occurring on the same haplotype (cis). Below that, there is a 3×3 table that contains the 9 possible combinations of genotypes for the two variants of interest. The number of individuals in gnomAD v2 that fall in each of these combinations are shown and are colored by whether they are consistent with variants falling on different haplotypes (red) or the same haplotype (blue), or whether they are indeterminate (purple). The estimated haplotype counts for the four possible haplotypes for the two variants as calculated by the EM algorithm is displayed on the bottom right. The probability of being in trans for this particular pair of variants is >99%. b, Variant co-occurrence tables on the gene landing page. For each gene (GBA1 shown), the top table lists the number of individuals carrying pairs of rare heterozygous variants by inferred phase, allele frequency (AF), and predicted functional consequence. The number of individuals with homozygous variants are tabulated in the same manner and presented as a comparison below. AF thresholds of ≤ 5%, ≤ 1%, and ≤ 0.5% are displayed across six predicted functional consequences (combinations of pLoF, various evidence strengths of predicted pathogenicity for missense variants, and synonymous variants). Both variants in the variant pair must be annotated with a consequence at least as severe as the consequence listed (that is, pLoF + strong missense also includes pLoF + pLoF).
