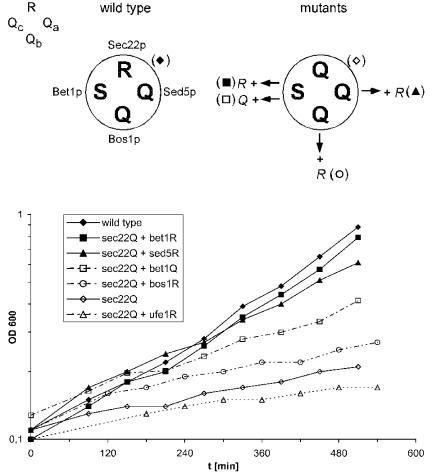
Figure 1.
Growth phenotypes of `0' layer Q→ R mutants analyzed for their ability to suppress the growth defect of the sec22(Q) mutation. The schematic drawings above the panel illustrate the arrangement of the amino acid side chains involved in the `0' layer of the ER-to-Golgi SNARE complex from wild-type and mutant yeast used in the growth assays. The ER-to-Golgi SNARE complex is composed of four proteins: R-SNARE Sec22p and the three Q-SNAREs Sed5p, a member of the syntaxin protein family, at position-Qa, Bos1p at Qb, and Bet1p at Qc. Note that, in contrast to mammalian Bet1 protein, yeast Bet1p carries a serine in the central layer. Bottom, growth curves of yeast cells (sec22Δ) expressing sec22(Q) as the sole version of SEC22 as well as the `0' layer mutants that were analyzed for their rescuing activity. The `0' layer mutants of SED5, BET1, BOS1, and UFE1 were expressed from plasmids in the presence of the normal copies on the chromosomes. Both bet1(R) and sed5(R) suppressed the growth defect caused by the sec22(Q) mutation to almost wild-type level, although the effect of the latter mutation rescued to a slightly lower extent. Yeast strain Y15177 (sec22::KanMX4) was transformed with the following sets of plasmids: pUA8-SEC22 and pRS316 (empty vector; filled diamonds), pUA4-sec22Q and pRS316 (open diamonds), pUA4 and pCG32-sed5R (filled triangles), pUA4 and pCG69-bos1R (open circles), pUA4 and pCG27-bet1R (filled squares), and pUA4 and pCG39-bet1Q (open squares). For control, strain Y15177 expressing pUA4-sec22Q was transformed with pCG5-ufe1R, coding for a `0' layer mutation in the syntaxin involved in retrograde transport to the ER (open triangles). Growth at 37°C was monitored by measuring the optical density at 600 nm. Selective minimal medium was used to prevent the loss of plasmids.