Figure 5. AR response elements are located near sex-biased genes.
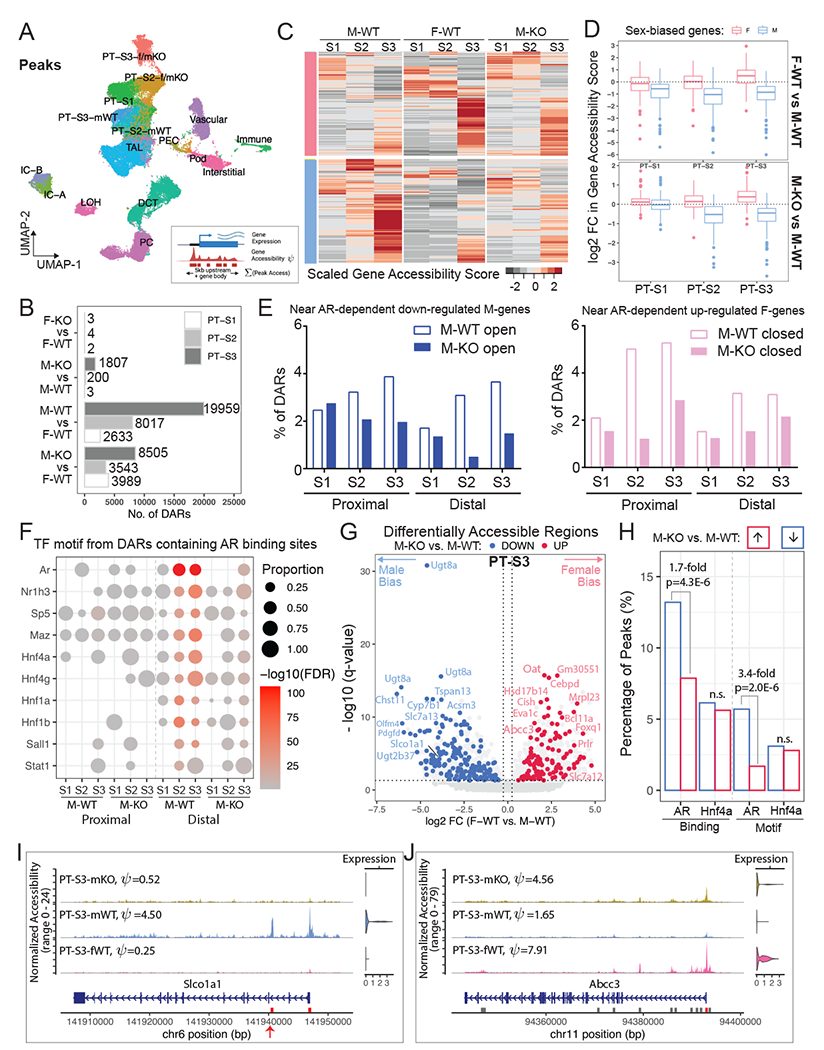
(A) UMAP plot shows clustering outcome using peaks called from the single-nuclear ATAC data. PT-S3-f/mKO: co-clustering of PT-S3 cells from F-WT, F-KO, and M-KO; PT-S2-f/mKO: co-clustering of PT-S2 cells from F-WT, F-KO, and M-KO; mWT: M-WT.
(B) Bar plot shows the number of sex-biased Differentially Accessible Regions (DARs) in PT segments identified for each pair-wise comparison (absolute Log2FC > 0.25, adjusted p-value < 0.05).
(C) Schematic summary of the computation of gene accessibility score ψ and heatmap shows the scaled gene accessibility score ψ for AR-responsive genes in M-WT, F-WT, and M-KO PT segments.
(D) Box plots demonstrate fold change in gene accessibility score ψ of AR-responsive genes within individual PT segments.
(E) Histograms showing the percentage of the proximal and distal DARs from M-WT and M-KO compared to F-WT that were nearby AR dependent down-regulated M-biased (blue) and up-regulated F-biased (pink) genes.
(F) Dot plot summarizes TF motif enrichment in the open DARs containing AR binding sites based on the published CHIP-seq dataset46. in M-WT and M-KO compared to F-WT PT segments.
(G) Volcano plot shows DARs within 100KB of sex-biased genes in PT-S3. 11,972 peaks were differentially open in male (left) and 7,987 peaks were differentially open in female (right). We colored F-biased peaks that are preferentially open in M-KO in red, and M-biased peaks that are preferentially closed in M-KO in blue. Each dot represents a 500-bp region, where the nearest gene is annotated.
(H) Bar plot shows the prevalence of TF binding and motif among PT-S3 sex-biased DARs that were altered by AR removal. TF binding was based on ChIP-seq data in the mouse kidney46. TF motif PWMs were retrieved from the Jasper database110 for AR (MA0007.3) and Hnf4a (MA0114.3).
(I-J) Coverage plots of two representative sex-biased genes, Slco1a1 (M-biased; I) and Abcc3 (F-biased; J). All peaks called in the region are shown in gray boxes, where DARs are highlighted in red. Peaks with potential AR binding site are indicated by red arrows.
