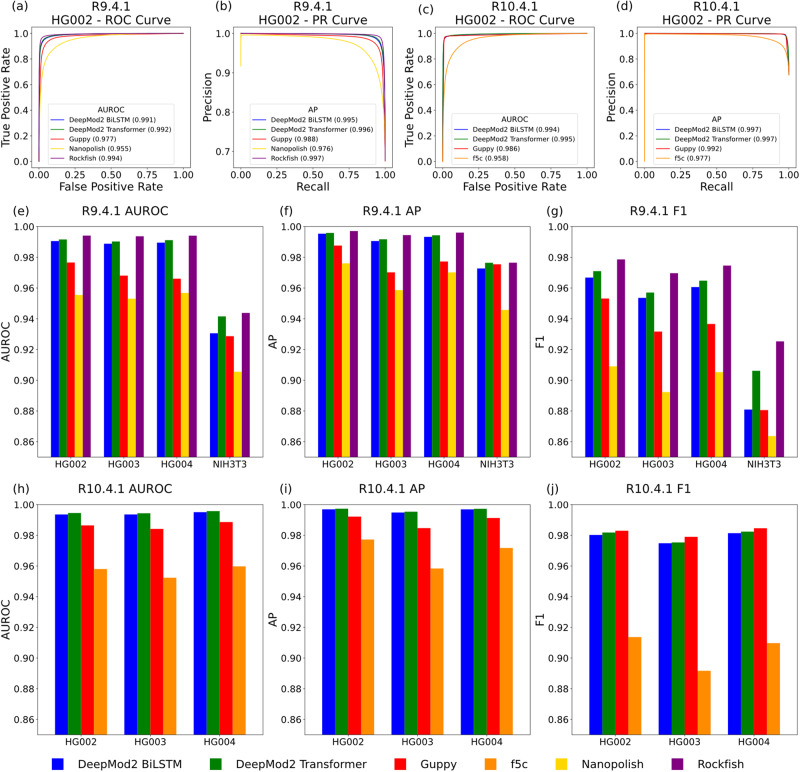
Fig. 3. Per-read performance of DeepMod2 BiLSTM and Transformer models and other state of the art methylation callers evaluated against short-reads ground truth for benchmarking.
a–d Show receiver operating characteristic (ROC) curves and precision-recall (PR) curves for R9.4.1 and R10.4.1 flowcell samples of HG002, with area under ROC (AUROC) and average precision (AP). e–g Show bar plots of AUROC, AP and F1-score for R9.4.1 flowcell datasets of HG002, HG003, HG004 and NIH3T3. h–j Show bar plots of AUROC, AP and F1-score for R10.4.1 flowcell datasets of HG002, HG003 and HG004. F1-score for each tool is evaluated at 0.5 probability of methylation threshold. Evaluation is performed on chr1 for HG002-4 and chr1-19 for NIH3T3. Source data are provided in the Source Data file.