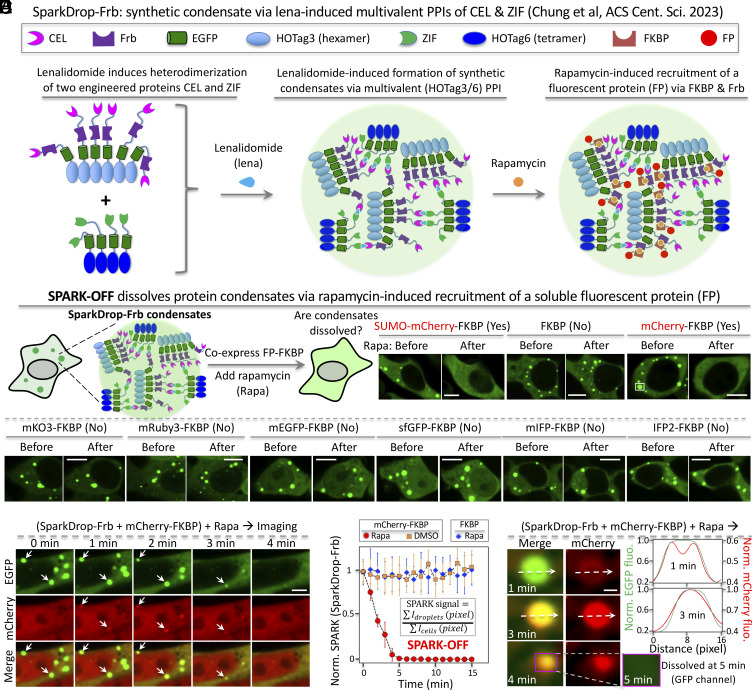
Fig. 4.
Designing a chemogenetic tool SPARK-OFF that dissolves protein condensates. (A) Schematic of synthetic condensates SparkDrop-Frb. (B) Schematic of SPARK-OFF and fluorescence images of HEK293 cells. The SparkDrop condensates are GFP-tagged synthetic condensates that form via multivalent interactions. Frb is tagged to the SparkDrop. The cells co-expressed FKBP-fused FPs. The cells were treated with rapamycin to induce the interaction between Frb and FKBP, resulting in recruitment of the FPs to the 20 SparkDrop condensates. SPARK-OFF uses mCherry to dissolve condensates. (C) Time-lapse images of HEK293 cells showing dissolution of SparkDrop condensates upon rapamycin-induced recruitment of mCherry. Arrows point to example condensates that are dissolved by SPARK-OFF. (D) Quantification of the condensate dissolution over time by rapamycin-activated SPARK-OFF by calculating SPARK signal that is defined by the ratio of the GFP in the condensates over total GFP. The SPARK signal at time 0 is normalized to 1. (n = 3 cells). (E) Time-lapse images of HEK293 cells showing mCherry recruitment to SparkDrop condensates and subsequent dissolution of the condensates. (Right) the fluorescence intensity profile along the position outlined in the images shown by the dashed lines. [Scale bar: 10 μm (B), 5 μm (C), 1 μm (E).]