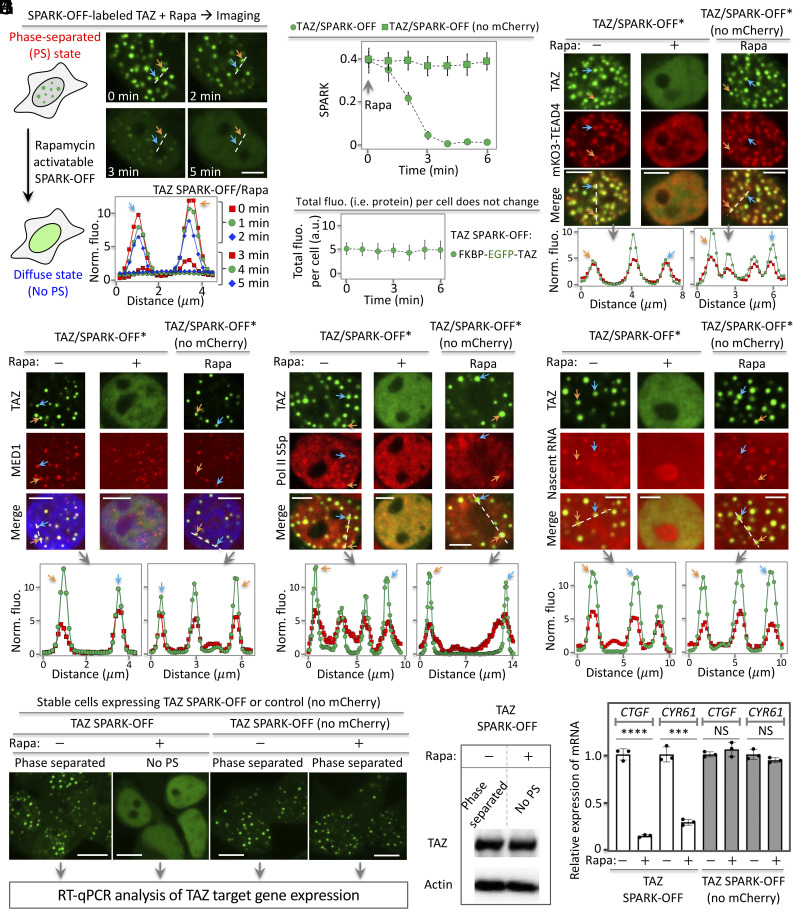
Fig. 5.
The chemogenetic tool SPARK-OFF reveals the role of TAZ PS on transcription. (A) Rapamycin-activatable SPARK-OFF dissolves TAZ condensates without change of protein level. The HEK293 cells expressed FKBP-EGFP-TAZ and NLS-mCherry-Frb. (B) Quantitative analysis of SPARK-OFF-driven dissolution of TAZ condensates over time by calculating SPARK signal over time. Data are mean ± SD (n = 3). (C) Total fluorescence of TAZ per cell over time upon SPARK-OFF. Data are mean ± SD (n = 3). (D–G) SPARK-OFF*-driven dissolution of TAZ condensates led to condensate dissolution of the of the DNA-binding and dimerization partner TEAD4 (D), transcriptional machinery including MED1 (E) and Pol II S5p (F), and nascent RNAs (G). SPARK-OFF* uses a non-fluorescent mCherry mutant (Y66F). (H) Fluorescent images of stable cells expressing SPARK-OFF-tagged TAZ or the control. The cells were treated with rapamycin or DMSO, followed by RT-qPCR analysis. No PS: no phase separation. (I) Western blot showing TAZ protein abundance level. (J) RT-qPCR analysis of the expression levels of two TAZ target genes in cells without and with PS of TAZ. Data are mean ± SD (n = 3). ****P-value < 0.0001, ***P-value < 0.001. NS, not significant. [Scale bars: 5 μm (A and D–G), 10 μm (H).]