Fig. 3. FDX1 starts the radical chain reaction of lipoyl biosynthesis.
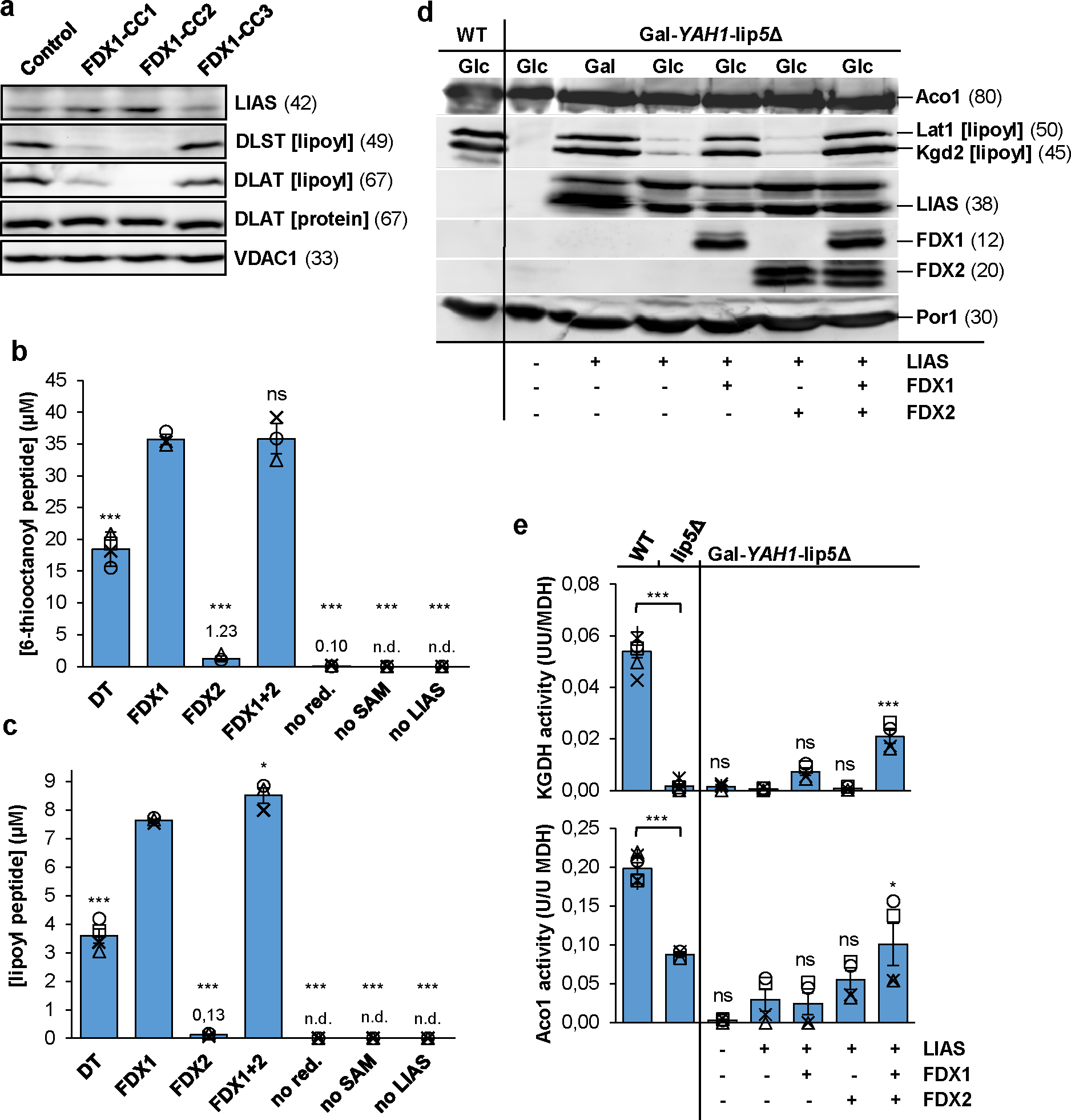
a HEK293 cell samples from Fig. 1b were immunostained for the indicated proteins or the lipoyl cofactor. VDAC1 served as loading control (see Fig. 1b). Observed molecular masses (kDa) for proteins are given in parentheses. Representative blots are shown. b, c Formation of the (b) 6-thiooctanoyl and (c) lipoyl peptide from a peptide substrate analogue [Glu-Ser-Val-(N6-octanoyl)Lys-Ala-Ala-Ser-Asp] by LIAS. Samples contained 0.5 mM peptide substrate, 35 μM LIAS, 2 mM NADPH, 20 μM FDXR, 140 μM FDX1, and 1 mM SAM, unless indicated otherwise. Dithionite (DT) samples lacked FDX1, FDXR, and NADPH. Reactions were incubated at 23°C, quenched with acid after 2.5 h, and products quantified by HPLC-MS using peptide standards (n ≥ 3; one-way ANOVA for FDX1 reactions versus all other conditions). d Gal-YAH1-lip5Δ yeast cells harboring plasmids encoding human LIAS, FDX1 and/or FDX2 as indicated were grown in minimal medium with glucose (Glc) or galactose (Gal) as indicated for 3 days. Mitochondria were isolated and analyzed for the indicated proteins or lipoyl cofactor in PDH and KGDH by immunostaining. Porin (Por1) was used as loading control. e Mitochondrial extracts were analyzed for aconitase (Aco1) and KGDH activities, and results were normalized to MDH activity. Wild-type (WT) and lip5Δ cells served as controls (n ≥ 4; one-way ANOVA, LIAS-complemented Gal-YAH1-lip5Δ cells versus all other Gal-YAH1-lip5Δ cells). *p < 0.05, ***p < 0.001. Error bars indicate the SEM. n.d., not detectable.
