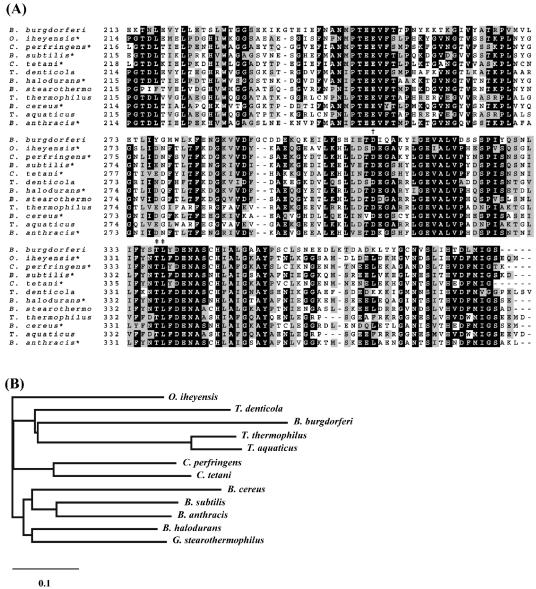
FIG. 4.
Sequence comparison and relationship of TAPBb to other members of the M29 family of metallopeptidases. (A) Multiple sequence alignment of C-terminal-portion amino acid sequences of aminopeptidases. The putative zinc binding (†) and bestatin ligation residues (‡) are represented. Sequences were obtained from the GenBank/EBI database under the following accession numbers: AAC66461 (Borrelia burgdorferi), BAC14998 (Oceanobacillus iheyensis), TDE2337 (Treponema denticola), P24828 (Geobacillus stearothermophilus), NP_389328 (Bacillus subtilis), NP_562913 (Clostridium perfringens), NP_782566 (Clostridium tetani), NP_243111 (Bacillus halodurans), NP_654259 (Bacillus anthracis), NP_831585 (Bacillus cereus), P42778 (Thermus thermophilus), and P23341 (Thermus aquaticus). (B) Phylogenetic relationship of members of the aminopeptidase M29 family. A phylogram was generated after alignment of the full-length amino acid sequences of the 11 enzymes, using CLUSTAL W software with a PAM250-weight table set with the parameters Ktuple = 1, opening penalty = 3, and gap extension = 5. The scale at the bottom represents the number of amino acid substitutions per site. Asterisks indicate functionally defined enzymes.