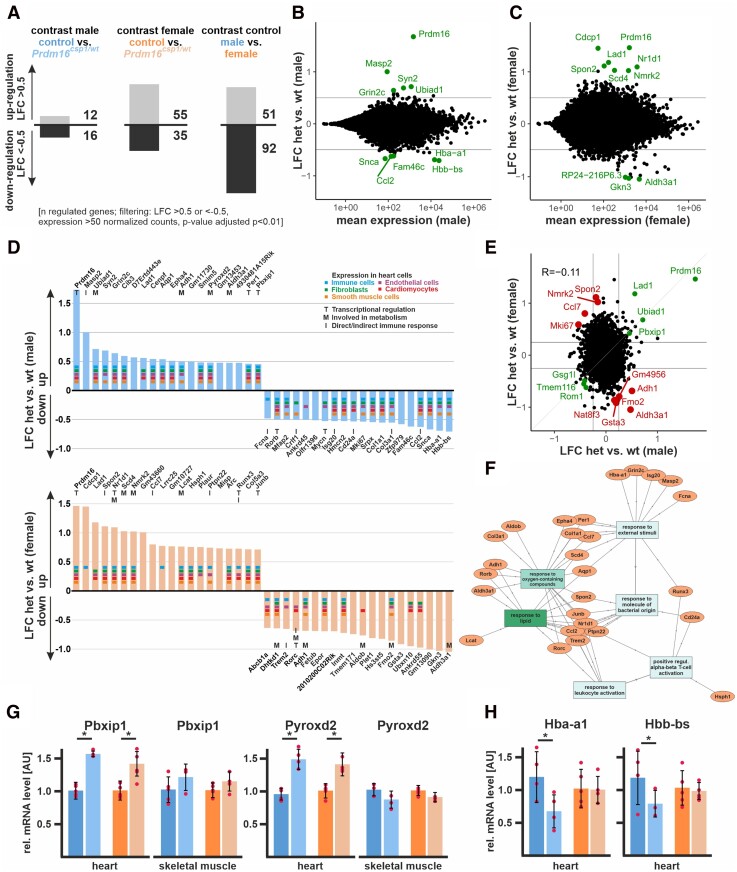
Figure 3.
Transcriptome analysis of Prdm16csp1/wt heart tissue. (A) Transcriptome analysis with RNAseq of Prdm16csp1/wt LV shows in male 28 and in female 90 differentially regulated genes compared to controls. Analysis of male vs. female control hearts identifies 143 differentially regulated transcripts. Analysed biological replicates are n = 4–6. (B and C) Bioinformatic filtering ranked the normalized log2 cpk according the absolute log2 fold change (LFC) after elimination of regulated targets with an adjusted P-value (padj) < 10−2. Scatterplots show the LFC against mean expression for the contrast Prdm16csp1/wt (het) vs. controls (wt) in male (B) or female (C) heart. The top 10 differentially regulated genes (adj. P < 0.01) are highlighted and labelled. (D) Summary of top 20 up- and down-regulated genes in male (upper panel) and female (lower panel) hearts. Expression of selected genes in cardiac cell types is annotated according to the colour code. Functional association with transcription (T), metabolism (M), and immune response (I) is shown for relevant genes. (E) Scatterplot shows the LFC detected in A and B against each other. Genes with concordant changes (abs LFC > 0.25) are highlighted in green, and genes with discordant changes (top 10; adj. P < 0.01) are highlighted in red. The overall correlation is R = −0.11 (n = 21 276). (F) Gene ontology (GO) network was constructed with GOnet18 by using the male and female TOP20 up- and down-regulated genes in Prdm16csp1/wt hearts. GO terms response to lipids and response to oxygen-containing compounds show strongest association. (G) Validation of RNAseq expression data for Pbxip1 and Pyroxd2 was performed with qPCR using whole RNA extracts from heart and skeletal muscle. Pbxip1 and Pyroxd2 expression is increased in hearts from Prdm16csp1/wt mice of both sexes. (H) Hba-a1 and Hbb-bs expression is diminished in male Prdm16csp1/wt hearts.