Fig. 3.
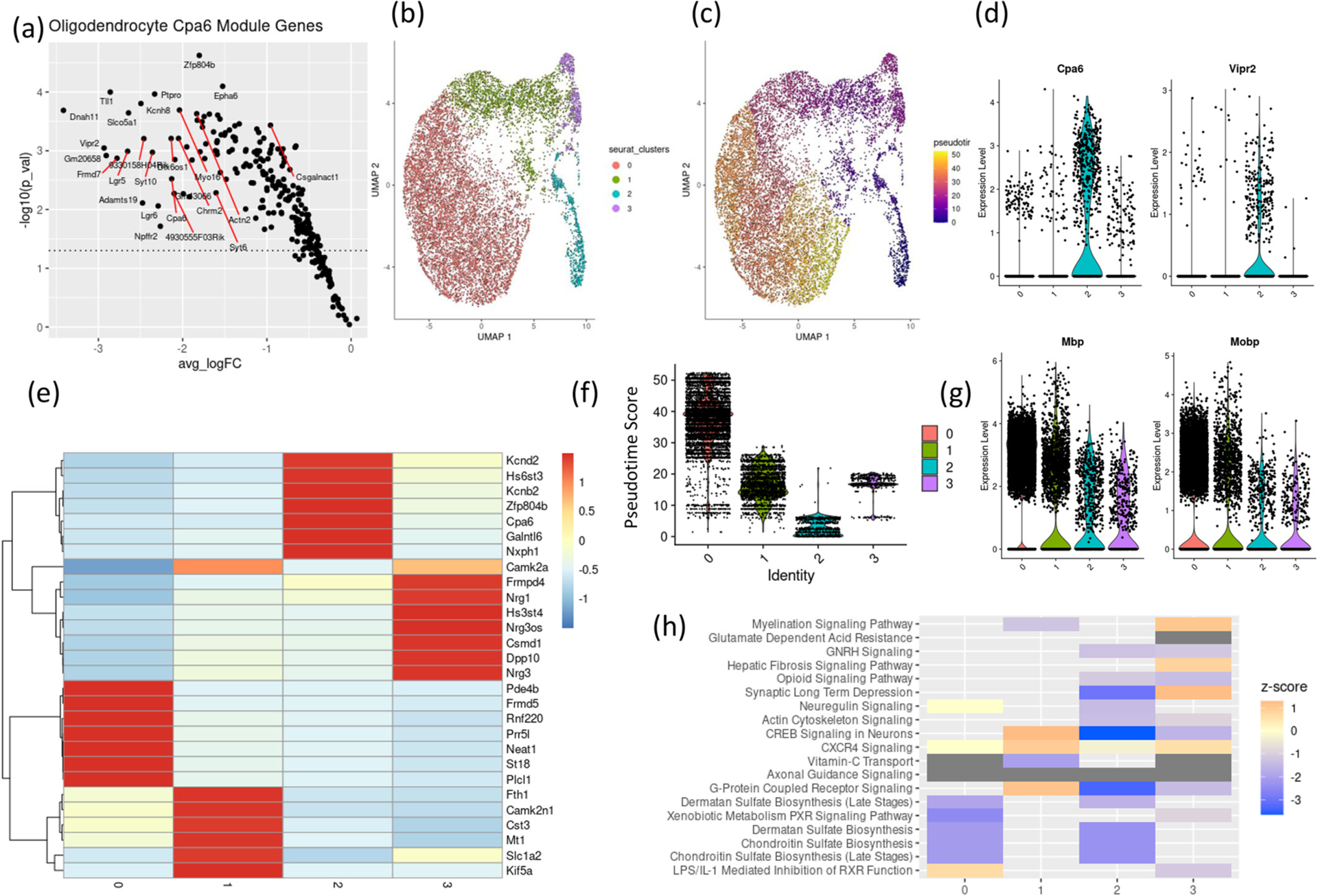
Oligodendrocytes sub clustering into transcriptionally distinct subtypes.
(a) Average log2 fold change (x-axis) and – log 10 (p-value) of gene members of oligodendrocytes Cpa6 module. UMAP plots of oligodendrocytes colored by (b) unsupervised clustering assignment and (c) pseudotime score.
(d) expression of Cpa6 and Vipr2 in oligodendrocyte subclusters.
(e) heatmap showing scaled expression values of seven top expressed genes in each oligodendrocyte subclusters.
(f) violin plots showing pseudotime scores in each oligodendrocyte clusters (g) expression of myelin related genes, Mbp & Mobp, in oligodendrocyte subclusters.
(h) z-score of select predicted pathways enriched in differentially expressed genes in oligodendrocytes subclusters, positive z-score = predicted activation, negative z-score = predicted inhibition.
