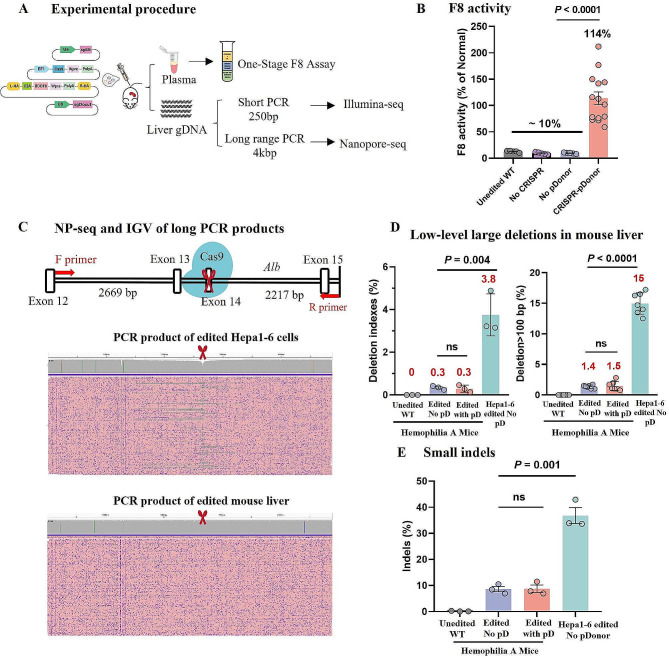
Fig. 1.
Evaluation of large deletions at the Alb target site in CRISPR and F8 donor gene-edited hemophilia A mice. (A) Schematic representation of the experimental workflow, involving hydrodynamic injection of editing plasmids into hemophilia A mice. Post-treatment assessments included measuring F8 activities via One-Stage F8 Assay and analyzing indels through Illumina sequencing, Crispresso2, and long-range PCR coupled with nanopore sequencing to detect potential large deletions. (B) Quantification of serum F8 activity three weeks post-administration in different groups: untreated mice (n = 5 mice), mice injected without CRISPR components (n = 5 mice), mice injected without pD-BDDF8-sg (n = 5 mice), and mice injected with both CRISPR components and pD-BDDF8-sg (n = 14 mice). Error bars represent mean ± SEM. (C) Using long-range PCR and nanopore sequencing to identify large deletions. The IGV visualization displays 200 randomly sampled reads, with purple dots marking sequencing errors and red scissors denoting the sgAlb target site. A positive control was established using Hepa1-6 cells. (D) Comparative analysis of large deletions in edited liver tissue from hemophilia A mice versus in vitro edited Hepa1-6 cells (n = 3), using deletion indexes and D100 (percentage of deletions > 100 bp). Statistical analysis was performed using unpaired two-sided Student’s t-tests. (E) Comparison of small indel frequencies in CRISPR-edited mice with or without pD-BDDF8-sg (n = 3 mice each) and in vitro edited Hepa1-6 cells (n = 3). Statistical evaluations were conducted using one-way ANOVA and unpaired two-sided Student’s t-tests