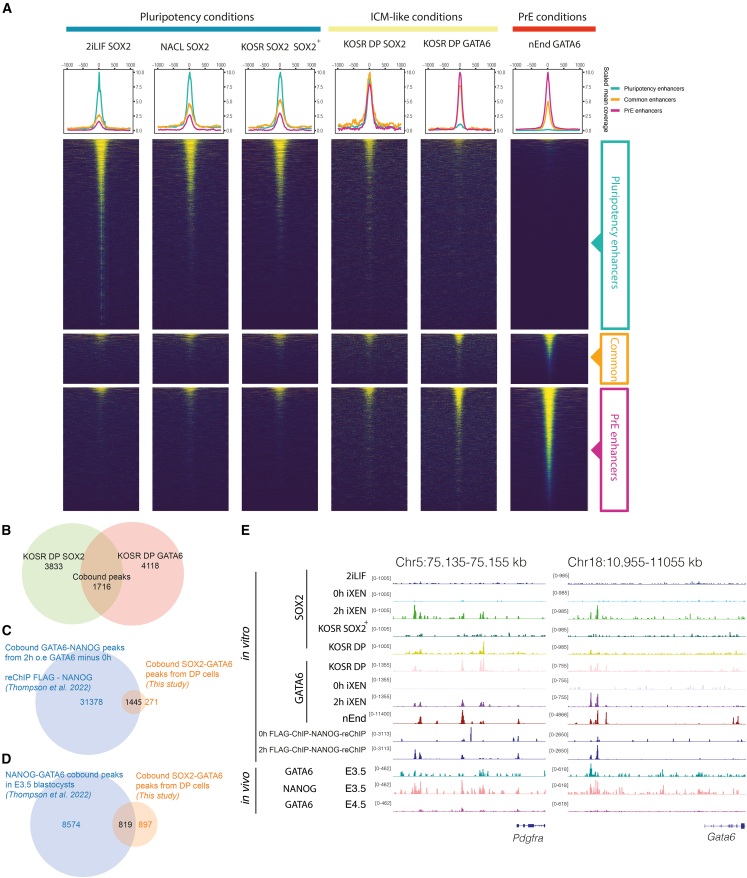
Figure 4.
Reciprocal lineage priming by GATA6 and SOX2
(A) Scaled heatmaps showing SOX2 and GATA6 binding at the pluripotency, common, or PrE enhancer subsets defined previously, in the conditions: 2iLIF, NACL, KOSR sorted cells, and nEnd.
(B) Euler diagram of SOX2 and GATA6 peaks in KOSR DP cells.
(C) Euler diagram showing the overlap of the GATA6-FLAG chromatin immunoprecipitation (ChIP) with NANOG reChIP (Thompson et al. 2022) with our SOX2-GATA6 co-bound peaks in DP cells.
(D) Euler diagram projecting the NANOG-GATA6 co-bound peaks from the in vivo data of the E3.5 blastocysts (Thompson et al. 2022) with the SOX2-GATA6 co-bound peaks in KOSR DP cells.
(E) Genome browser tracks of SOX2 and GATA6 comparing data from this study (2iLIF, KOSR, and nEnd tracks) with Thompson et al. (2022) (0 h and 2 h of iXEN induction, the GATA-FLAG ChIP with NANOG reChIP and the in vivo E3.5 and E4.5 blastocysts tracks). Pdgfra and Gata6 loci. Tracks are scaled accordingly with antibody and experiment. All the in vitro SOX2 experiments are in the same scale, the in vitro GATA6 experiments are also in the same scale (apart from nEnd due to the extreme difference in PrE genes expression). FLAG-NANOG reChIP tracks are on the same scale, as well as the in vivo data.