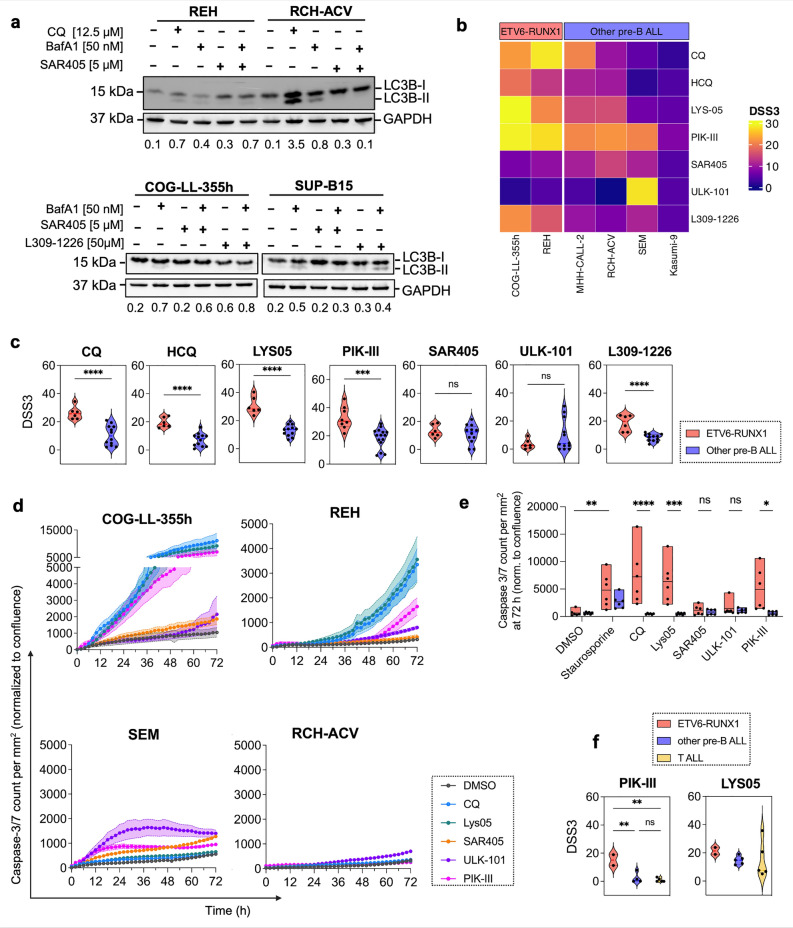
Figure 2.
ETV6-RUNX1 ALL cells are particularly sensitive to autophagy inhibitors. (a, upper panel—one membrane) REH and RCH-ACV cells were treated with either CQ or SAR405 for 24 h in the presence or absence of BafA1 and LC3B-II levels were monitored by Western blotting. (a, lower panel) COG-LL-355 and SUP-B15 cells were treated with L309-1226 or SAR405 for 24 h in the presence or absence of BafA1 and LC3B-II levels were monitored by Western blotting on two separate membranes for each cell line. Representative image of three independent experiments is shown. GAPDH is used as a loading control. Each membrane that was cut after transfer according to the corresponding protein sizes. Ratio of LC3B-II to GAPDH is quantified using Image J and presented under the figure. (b, c) Indicated cell lines were treated with indicated autophagy and lysosomal inhibitors for 72 h and viability was assessed using CellTiter-Glo. DSS3 was calculated using DSRT online tool; higher DSS3 values indicate higher sensitivity. Data is presented as a heatmap (b) or violin plots (c) of individual experiments comparing the two ETV6/RUNX1 to four other pre-B-ALL cell lines (n ≥ 3). (d, e) IncuCyte live-cell imaging of cells treated with the indicated autophagy inhibitors for 72 h (n = 3). (d) caspase 3/7 activity is normalized to cell confluence and presented over time. Note the different Y-axis scale. Treatment with 0.1 µM Staurosporine was used as a positive control for apoptosis induction. (e) Quantification of mean relative caspase 3/7 levels at the 72 h time point ± SEM (n = 3, each performed in triplicate). (f) Primary ALL cells were cultured ex vivo and treated with indicated autophagy/lysosomal inhibitors in either 96 or 384-well plates for 48 h (n = 1 for 11 individual samples, each performed in triplicate). Cell viability was measured using CellTiter-Glo and drug sensitivity scores were generated using the DSRT online tool; higher DSS3 values indicate higher sensitivity. * p < 0.05; ** p < 0.01; *** p < 0.001, **** p < 0.0001 using either a two tailed t-test with Welch’s correction for two groups (c) or 2-way ANOVA with Šidák (d) or Tukey (f) multiple comparison tests.