Figure 1: The AFRC is a pre-parameterized polymer model based on residue-specific polypeptide behavior.
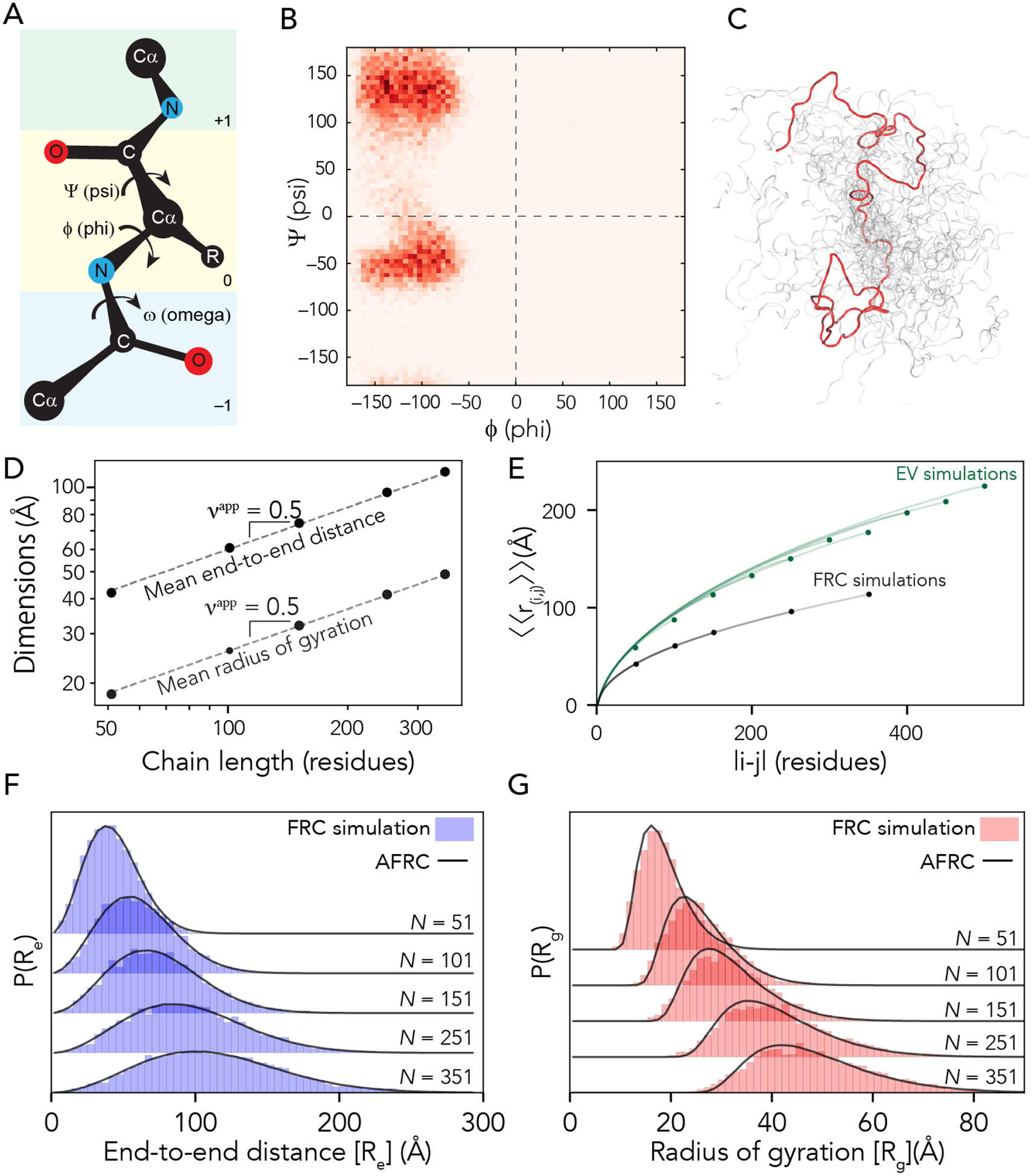
A. Schematic of the amino acid dihedral angles. B. Ramachandran map for alanine used to select acceptable backbone conformations for the FRC simulations. All twenty amino acids are shown in Fig. S1. C. Graphical rendering of an FRC ensemble for a 100-residue homopolymer. The red chain is a highlighted single conformation, and the shaded other chains shown to highlight the heterogeneous nature of the underlying ensemble. D. Flory Random Coil (FRC) simulations performed using a modified version of the ABSINTH implicit model and CAMPARI simulation engine yield ensembles that scale as ideal chains (i.e., Re and Rg scale with the number of monomers to the power of 0.5). E. Internal scaling profiles for FRC simulations and Excluded Volume (EV) simulations for poly-alanine chains of varying lengths (filled circles demark the end of profiles for different polymer lengths). Internal scaling profiles map the average distance between all pairs of residues |i-j| apart in sequence space, where i and j define two residues. This double average reports on the fact we average over both all pairs of residues that are |i-j| apart and do so over all possible configurations. EV simulations show a characteristic tapering (“dangling end” effect) for large values of |i-j|. All FRC simulation profiles superimpose on top of one another, reflecting the absence of finite chain effects. F. Histograms of end-to-end distances (blue) taken from FRC simulations vs. corresponding probability density profiles generated by the Analytical FRC (AFRC) model (black line) show excellent agreement. G. Histograms of radii of gyration (red) taken from FRC simulations vs. corresponding probability density profiles generated by the AFRC model (black line) also show excellent agreement.
